Team:Heidelberg/Templates/Delftibactin Results
From 2013.igem.org
Contents |
Results
Producing Delftibactin
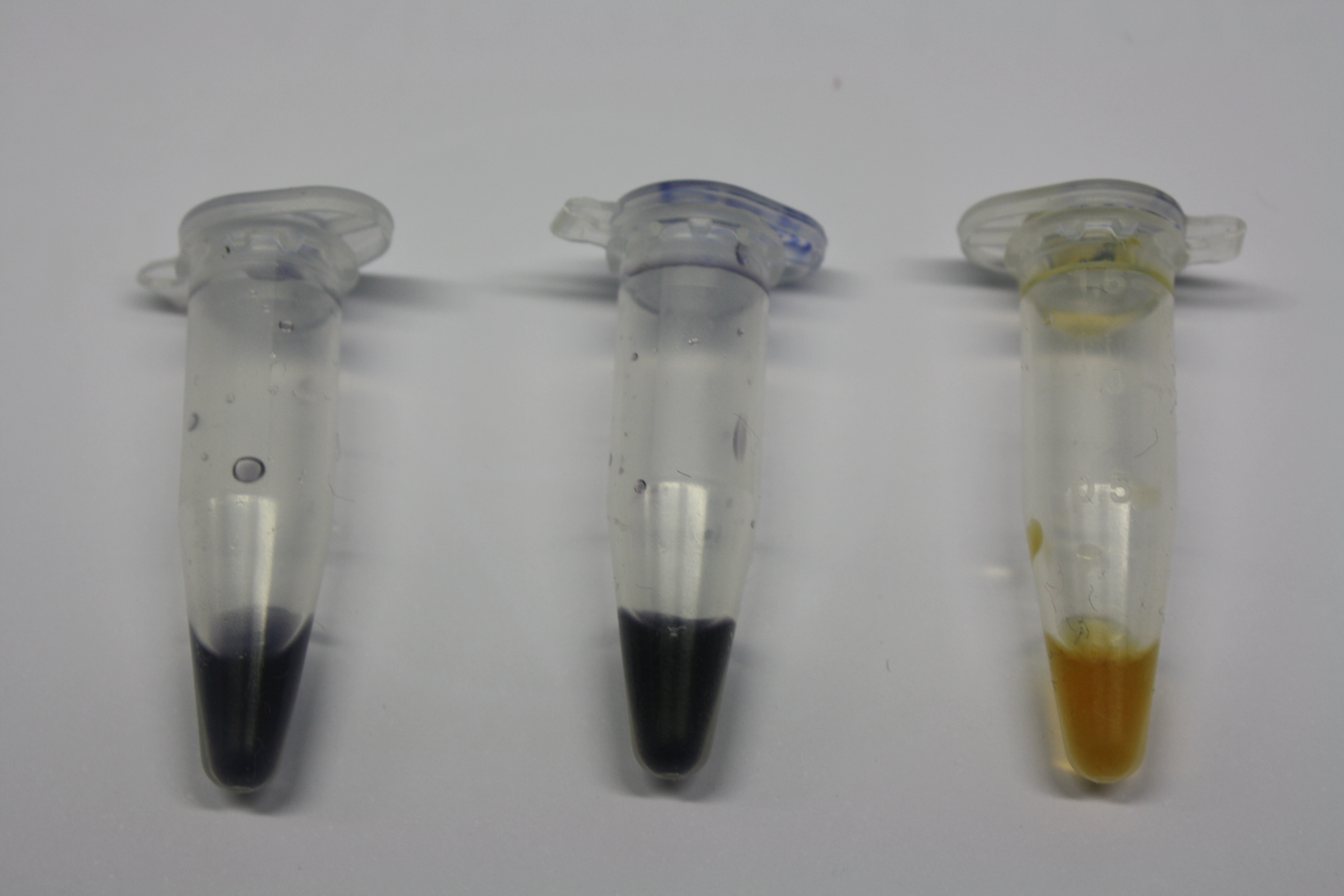
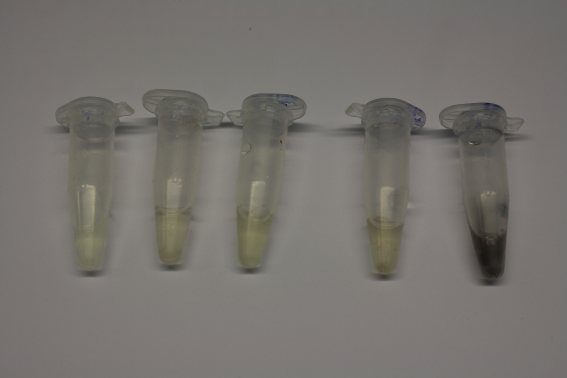
We obtained D. acidovorans DSM-39 from the ZSMZ and successfully reproduced the paper of Johnsson et al. (<bib id="pmid23377039"/>). In our experiments, precipitation on agar plates worked even better than described in the paper as shown in Figure 1. D. acidovorans is capable to precipitate solid gold from gold chloride solution as purple-black nanoparticles. Already at low concentrations of gold chloride, gold nonaparticles are precipitated increasing with concentration of gold chloride in solution (Fig. 2).
Using supernatants from the new Delftia acidovorans strain SPH-1, we showed precipitation of gold chloride solution to gold nanoparticles. Furthermore, we melted the purple-black nanoparticles to shiny solid gold as shown in figures 3 to 5.
We established purification of Delftibactin using HP20 resins. Additionally, we proved precipitation of gold by the purified Delftibactin (figures 6 and 7) and detected it by Micro-TOF File:20130911Malditof.pdf.
On the way to biological recycling by Delftibactin, we were able to dissolve gold-containing parts of an old CPU and established a protocol for recovery of gold as soluble gold salts from electronic waste (figures 8 to 10).
Amplification of the Del Cluster Genes
The first step towards introducing the Delftibactin pathway into E. coli was the amplification of the Del-cluster encoded on the genome of Delftia acidovorans. To this end, we designed Gibson primers and amplified the genes of the non-ribosomal peptide synthetase and the polyketide synthase (PKS) pathway as well as additional constitutive proteins, which were predicted to be necessary for the production of Delftibactin [1] . In the first weeks, PCRs were successfully established and optimized. At the same time, a separate plasmid was created encoding the PPTase from Bacillus subtilis and a permeability device [ BBa_I746200] [2]for the export of the synthesized NRP. Additionallly, plasmids from the partsregistry [3] were successfully transformed and amplified for the backbone generation.
Gibson Assembly & Transformation
As it is not trivial to assemble such a large number of long DNA fragments, we have used the Gibson Assembly method [4] implemented by Cambridge in iGEM 2010 instead of common cloning procedures such as restriction and ligation techniques. The assembled constructs of up to 32 kbp in size were transformed into E. coli via electroporation. Correct assembly of the fragments was tested by analytical restriction digest.
Figure 11: Restriction digest of three3 different plasmids needed for the NRPS/PKS pathway which to generated Delftibactin. a) Four4 digested colonies clones of the pIK8-plasmid, where clones 6 and 9 show the expected pattern. b) shows oneOne clone of E. coli clone with the 32 kpb DelRest -plasmid and was digested with three different enzymes and every lane shows the specific pattern for the according enzyme. c) Rshows the restriction digfest of the DelH -plasmid with PvuI. Clone 5 shows the expected pattern and is probably positive.
The exemplary restriction digest shown above (Fig. 11) confirms the correct assembly of the three desired constructs as it shows the expected band pattern expected from in silico digestion. Clones (6 and 9) contained the plasmid that encodes for the Methylmalonyl-CoA pathway (Fig. 11a) .The obtained DNA sequences were sent in for sequencing over the gibson-assembled regions for confirmation. [5][6] The sequencing confirmed the accuracy of the sequence. The successful generation of DelRest plasmid was proven by different enzymatic restriction digests (Fig. 11b) and also attested by the sequencing. The sequence was compared with the available SPH1 sequence of the Del cluster of Delftia acidovorans in NEB-database [7] (For further information please visit our labjournal).
After analytical restriction digest of DelH (Fig.11c), dozens of clones were positive. In contrary to the two plasmids before, the sequences prepped from the DelH clones exhibited a variation of mutations. Most of them being deletions at the very beginning of the DelH`s coding region. Interestingly, one specific deletion in the DNA sequence was found consistently in several independent clones. As we did not have any clone without mutations, we proceeded with the a DelH clone (termed C5), as it did not have any DNA deletion but that only harbored a minor base- pair substitution was observed not leading to a frame shift but a conversion from Alanine to Threonine at the N-terminus of the protein. Some exemplary sequences are listed below (Tab. 1) of Delftia acidovorans and two observed mutations in different DelH-clones.
Tab.1 DNA sequences of DelH from the start codon. The table shows the sequence Comparison between template strand of D.acidovorans naturally expressing hosting the NRPS/PKS pathway and two different exemplary clones of E. coli transformed with the plasmid pHM04. The plasmid was Gibson-assembled to contain the DelH gene. The second line shows the accumulated deletion and the third line shows the clone containing 'only' a substitution. Deletions appeared quite frequently while a substitution was only found in clone C5. The substitution changes the Alanine codon to Threonine.
| Organism | Plasmid containing | DNA -Sequence | Conclusion |
|---|---|---|---|
| D. acidovorans | ATG GACCGTGGC CGCCTGCGC CAAATCGC | correct | |
| E. coli DH10ß | ATG GACCGTG-C CGCCTGCGC CAAATCGC | deletion | |
| E. coli DH10ß C5 | ATG GACCGTGGC CGCCTGCGC CAAATCAC | substitution |
After being proven to be potentially correct the above named constructs were used for further experiments.
The pictures in Fig. 11 show identification of potentially correct constructs via analytic restriction digests (Fig. 1), thus demonstrating suitability of the Gibson assembly technique to clone DNA constructs. Furthermore, we successfully transformed all three plasmids into E. coli DH10ß or E. coli BL21 DE3, respectively.
Due to the fact that E. coli seemed to somehow selected for mutated DelH clones, we hypothesize that expression of DelH in absence of the other Del cluster proteins be toxic for the cells .This would explain why E. coli selects for mutated, none functional, truncated DelH proteins. The same phenomenon of frequent mutations in presumably positive clones was also observed for cloning of the permeability device used in the pIK8 construct. The sequenced plasmids showed an unusually high accumulation of mutations compared to other constructs. In case of the methylmalonyl-CoA plasmid (pIK8), the problem was solved by the usage of a weak promotor and a weak ribosome binding site from the partsregistry. During the establishment of the methylmalonyl-CoA pathway cloning we identified a weak promotor and ribosome binding site to significantly reduce the expression of possibly toxic effects of the target and decreased selection pressure for mutations. Based on this knowledge, DelH is currently being assembled into a new backbone (pSB6A1) [8] containing the promotor BBa_J23114 [9] and the ribosome binding site BBa_B0032[10]. While the new plasmid is constructed, the following experiments were performed with the C5 clone (harboring only an amino acid substitution Tab.1) for expression control.
Expression of the Delftibactin NRPS & Associated Genes
We carried out experiments to test whether the NRPS/PKS pathway, together with the PPTase, the Methylmalonyl-CoA pathway and the permeability device are all efficiently expressed after transformation into E. coli. The transformation was performed as well by electroporation. Different approaches were tested, including triple-electroporation and as well as double-electroporation with an additional single transformation afterwards (testing all combinations). There were no significant differences observed.
To for expression of DelH and DelRest, we conducted SDS-PAGEs. As negative controls we used untransformed cells and cells transformed only with the original backbone .The proteins DelE, DelG and DelH are significantly larger than any protein that is expressed by our host E. coli. Therefore, the expression of the introduced genes was clearly visible on the SDS-PAGE (Fig. 12). Even though the expression was weak, as we have expected for such a large protein, clear distinct bands at the expected size of DelE and DelG can be detected in the clone transformed with the DelRest plasmid and a band at the size of DelH for the clone transformed with the DelH plasmid. As the promotor in front of DelE and DelG controls the expression of DelA, DelB, DelC, DelD and DelF, too, one can assume simultaneous expression of these Del proteins.
Furthermore, the expression of the PPTase was verified by co-transformation of the referring plasmid pIK8 with an indigoidine synthetase. The transformed E. coli grew very slowly and but developed a clear blue color. From these results, we can clearly conclude that the PPppTase is functionally expressed: The indigoidine synthetase requires the presence of a functional PPTase, as it is needed for the activation of the indigoidine synthetase, which then finally produces the blue pigment indigoidine. Decelerated growth kinetics of E. coli points at the metabolic burden that is implied by the synthesis of the indigoidine.
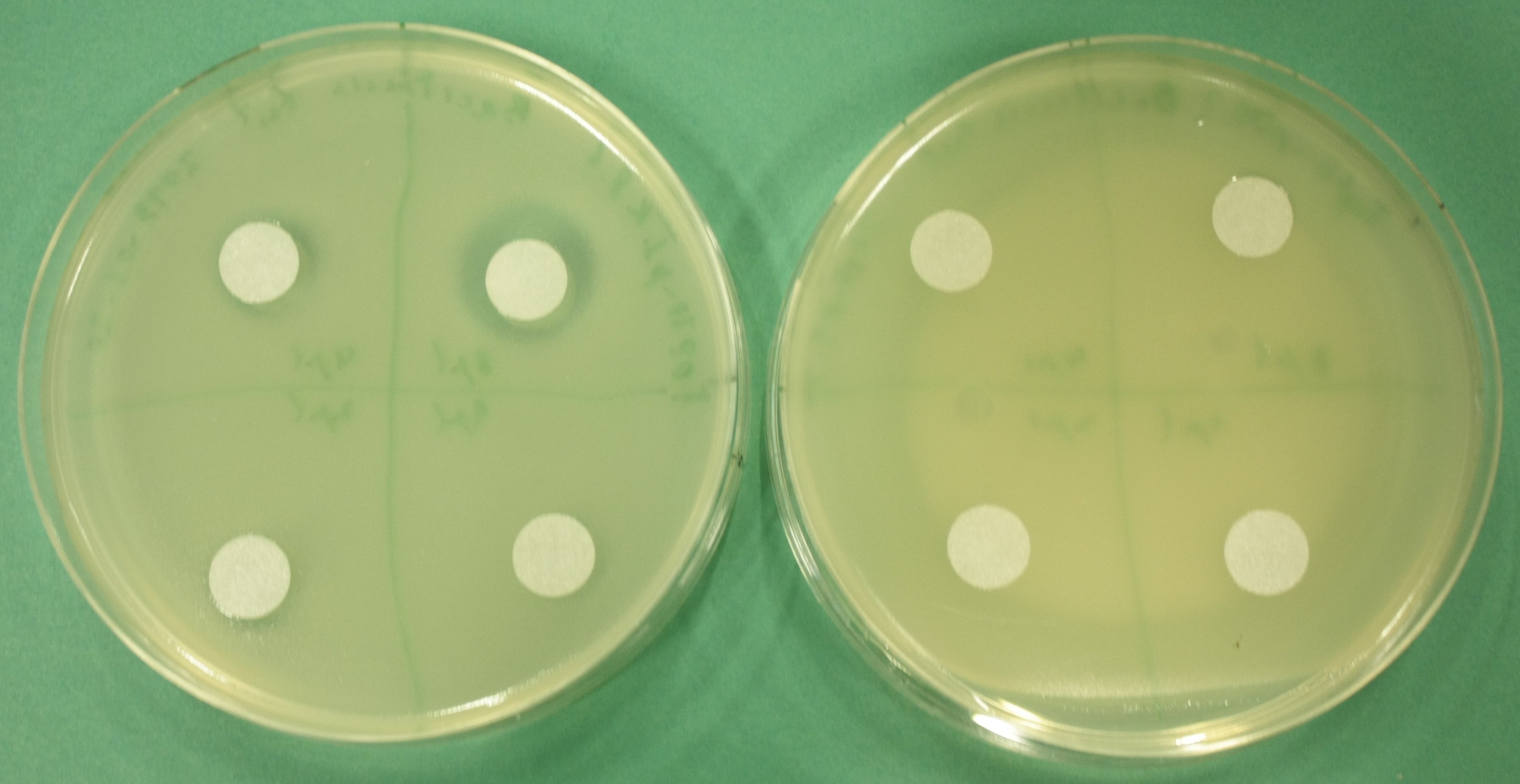
For proving that E. coli also produces the permeability device, which is needed demanded to export Delftibactin out of thethe cells, a Hemmhof agar diffusion test with bactracin was performed. Bacitracin is a very large antibiotic which that is usually not able to diffuse across the cell membrane passively. Absent growth upon application of bacitracin of bacteria containing the plasmid while in the control cells without the device were not affected by the antibiotic (Fig. 13) confirms expression of the transporter.
In conclusion, we successfully expressed the recombinant Delftibactin NRPS/ PKS pathway as well as the required Methylmalonyl-CoA pathway, the PPTase and permeability device . It is not only possible to assemble large plasmids (in sum up to 64 kpb) and transform them into E. coli, but also express NRPS modules and constitutive proteins.
 "
"