Team:Heidelberg/Templates/Indigoidine week19
From 2013.igem.org
pRB15-18
The first mini
Screening delC
| Template | Primer | Comment |
|---|---|---|
| TOP10+pSB3K3 | RB64/VR | negative control |
| VF2/65 | ||
| TOP10+pRB18 | RB64/VR | screening |
| VF2/65 | ||
| VF2/VR | ||
| delC PCR product | RB64/RB65 | positive control |
Heidelberg 20130902 screening
|
Screening pKH4
| Template | Primer |
|---|---|
| TOP10+pKH4 | KH52/VR |
| VF2/RB71 |
Heidelberg 20130902 screening
|
A miniprep of probe 4 was sent to sequencing, which was positive.
Assembly of pKH4-derivates
pKH4 is used for assembly of pKH5 and variants of pKH4 with all the different T-Domains.
| KH3/4 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| td 62 | 5 | |
| 72 | 2.00 | |
| 25 | 98 | 1 |
| 57 | 5 | |
| 72 | 2.00 | |
| 1 | 72 | 6.00 |
| 1 | 12 | - |
Heidelberg 20130902
|
KH3/4-PCRs were unsuccessful, so we did a gradient PCR
| KH3/4 in biometra | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 58-59-60-61-62-63 | 5 | |
| 72 | 2.00 | |
| 1 | 72 | 6.00 |
| 1 | 12 | - |
Heidelberg 20130903 KH3-4 gradient.png
|
We run the same protocol for 3x 50 ul.
Heidelberg 20130903 KH3-4.png
|
nothing
16 x 10 ul with Q5 and Tm calculator suggested parameters (63 °C annealing), pRB19 ligation as template
Heidelberg 20130903 KH3-4 16x 10
|
We tried NEB Phusion HF with 61 °C annealing temperature in a 10-, 20- and 50 ul reaction from pRB19 ligation and in a 20 ul reaction from pKH4 ind T100 (left). As well we ran with Phusion HF (59 °C annealing) in 20 ul reactions with 0, 1 and 2 ul DMSO (Biorad Cycler 2)
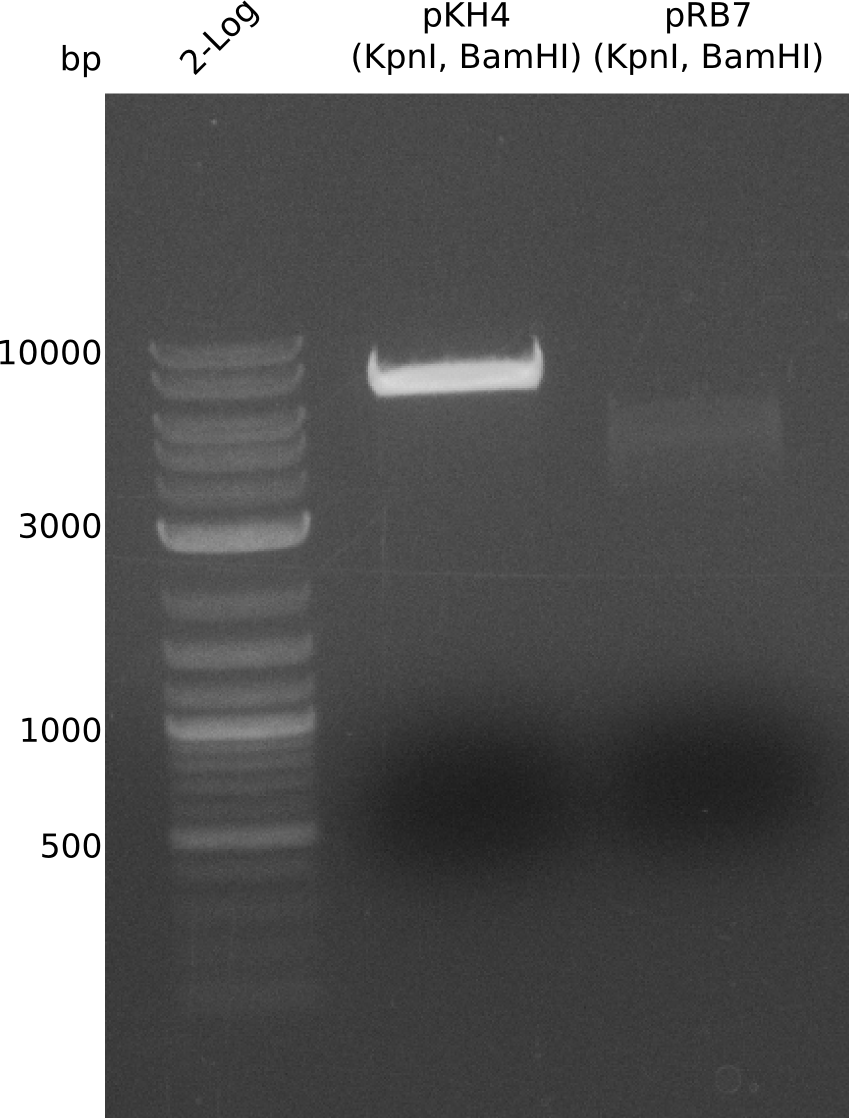
For further evaluation of the single plasmid strategy we admired a plasmid with bpsA instead of indC as indigoidine synthetase. For a quick shot we wanted to use pKH4 and pRB7 for a restriction approach with the enzymes KpnI and BamHI.
- prepare digestion (20 µl), 37 °C, ~0.5 h:
- pKH4: 2 µl NEBuffer CutSmart, 4 µl pKH4, 13 µl H2O, 0.5 µl (KpnI, BamHI)
- pRB7: 2 µl NEBuffer CutSmart, 2.3 µl pRB7, 14.7 µl H2O, 0.5 µl (KpnI, BamHI)
The restriction for pKH4 did not work correctly due to the fact that with assembly of pKH4 the BamHI cutting site was removed and the plasmid was only linearized by the KpnI cutting site. For pRB7 two bands at almost equal site can be expected and thereby the digestion approach can not be used in this manner. We will try again with a similar approach like for the assembly of pKH4 as here a nonsense sequence instead of the PPtases will be introduced into pRB7.
- preparation of pRB7 MP by retransformation of TOP10 (as no glycerol stock is available).
Amplification of svp
The svp fragment we used so far was wrong, so we try to amplify it again from the genome, using various primers:
- RB13/14
- RB15/16
- RB29/30
- RB39/40
Gradient on biometra cycler (72 °C, 65 °C, 58 °C)
Heidelberg 20130902 screening
|
svp was always right; only after assembly it turned wrong. We will try to bring it in using restriction cloning and to sequence it; maybe the screening is just inconclusive.
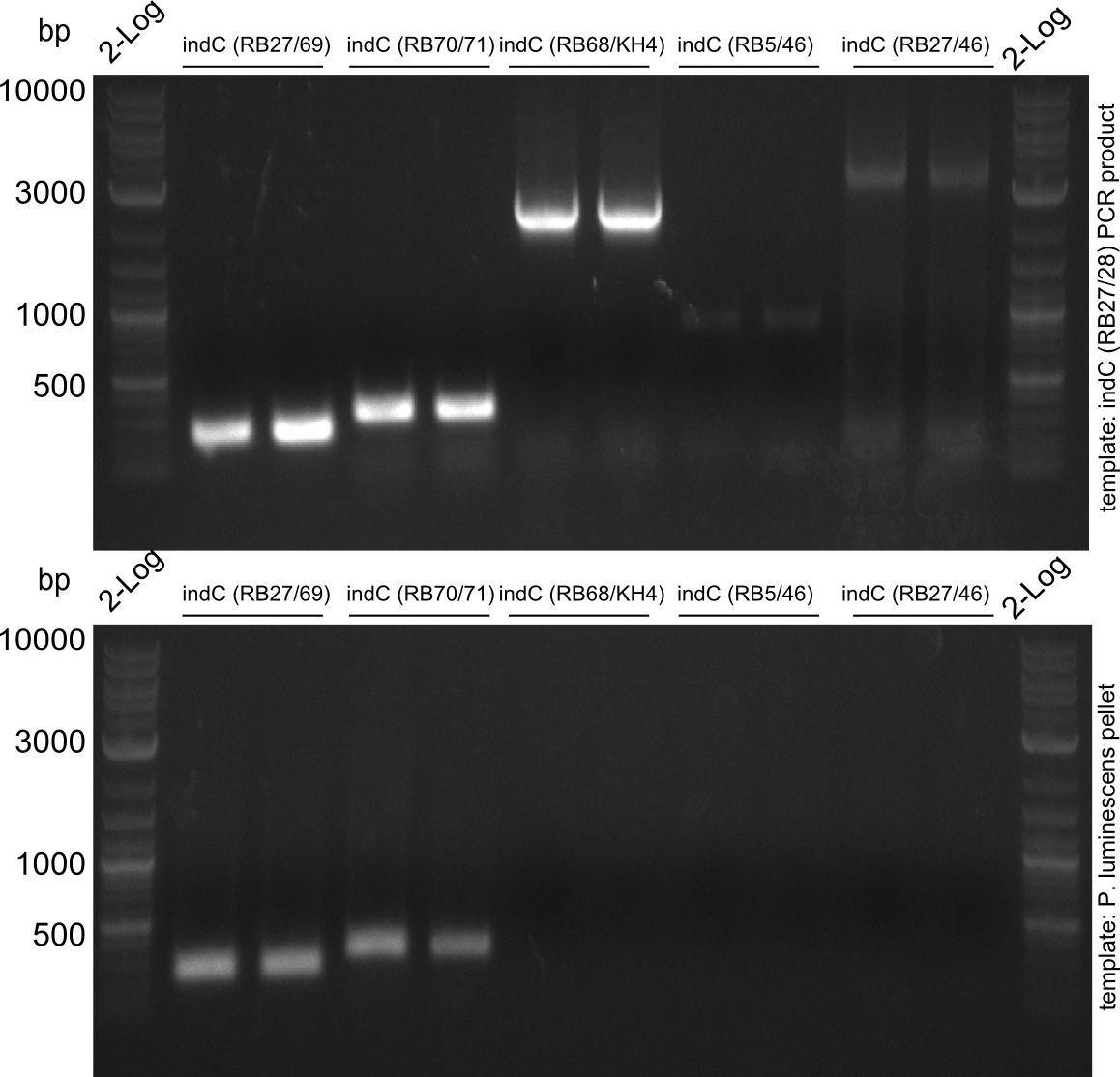
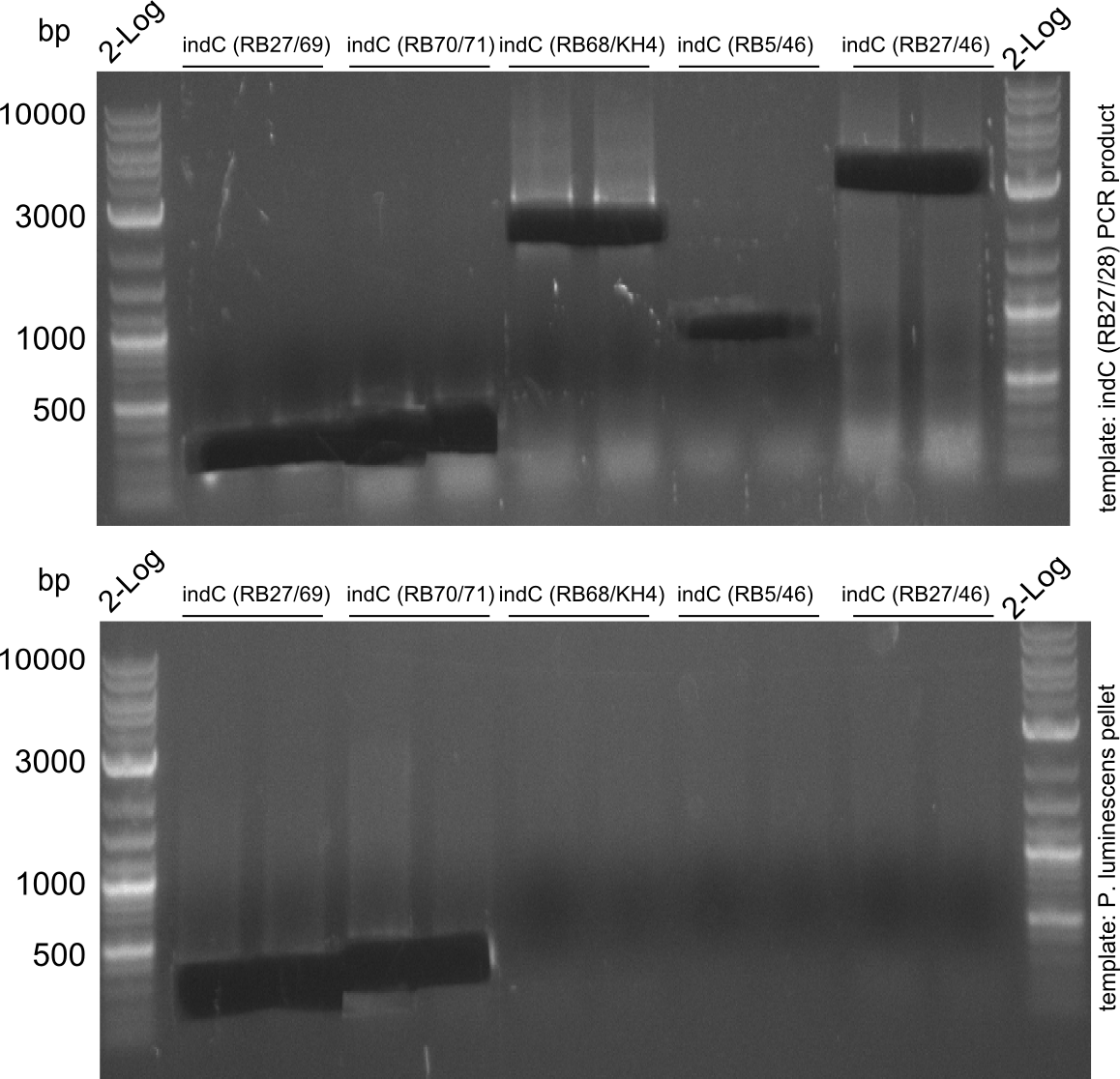
Amplification of indC-fragments
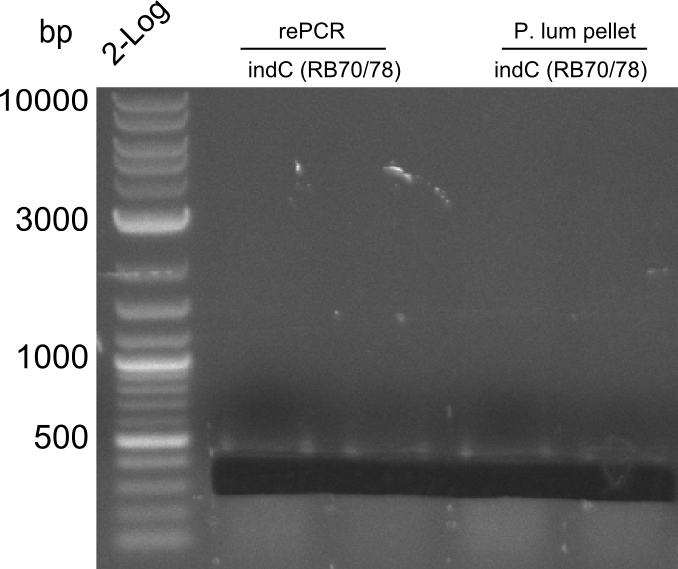
indC fragments are amplified from Plum genome and indC short PCR product in a 50 ul Phusioin HF reaction. The fragments are:
- RB27/69
- RB70/71
- RB68/46
- RB68/KH4
- KH5/RB46
- RB27/46 from pRB14
- RB27/28
| RB27/69 in BioRAD T100 from indC short | ||
|---|---|---|
| RB27/69 in BioRAD T100 from P. luminescens pellet | ||
| RB70/71 in BioRAD T100 from indC short | ||
| RB70/71 in BioRAD T100 from P. luminescens pellet | ||
| KH5/RB46 in BioRAD T100 from indC short | ||
| KH5/RB46 in BioRAD T100 from P. luminescens pellet | ||
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 2.00 |
| 35 | 98 | 5 |
| 55 | 15 | |
| 72 | 30 | |
| 1 | 72 | 2.00 |
| 1 | 12 | - |
| RB68/46 in BioRAD T100 from indC short | ||
| RB68/46 in BioRAD T100 from P. luminescens pellet | ||
| RB68/KH4 in BioRAD T100 from indC short | ||
| RB68/KH4 in BioRAD T100 from P. luminescens pellet | ||
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 2.00 |
| 30 | 98 | 15 |
| 55 | 15 | |
| 72 | 2.00 | |
| 1 | 72 | 5.00 |
| 1 | 12 | - |
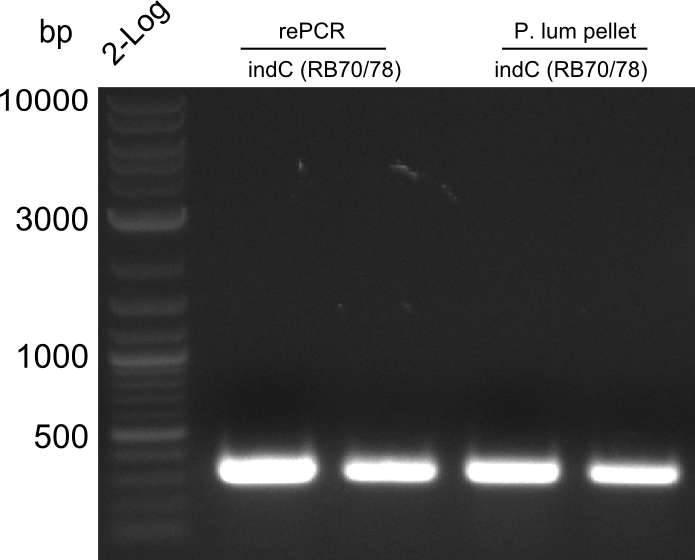
Fragment RB70/71 is wrong because RB71 is wrong. We repeated the amplification with RB78 instead of RB71.
Fragment KH3/RB46 is very weak, so we try to improve this reaction as well.
| KH3/RB46 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| td 60 | 5 | |
| 72 | 20 | |
| 23 | 98 | 1 |
| 63 | 5 | |
| 72 | 20 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
And reaction RB68/RB46 with the same protocol but 60 s elongation time.
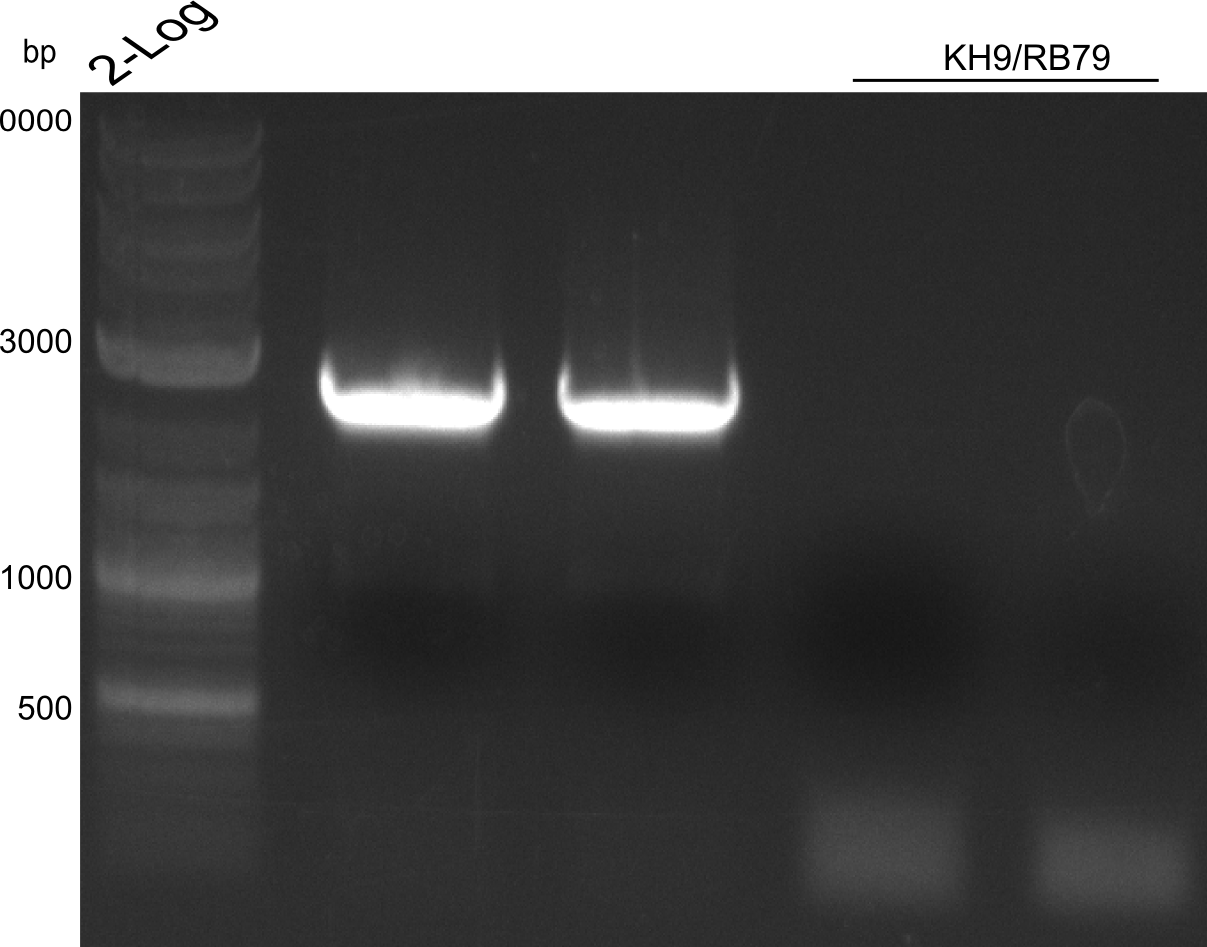
We try KH3/RB79 with improved conditions.
We tried to get this fragment with KH9/RB79 with Q5 in biometra cycler (60 °C annealing)
We tried to get this fragment with KH9/RB79 with Phusion HF in biometra old cycler (61 °C annealing)
Gel Extraction yielded xx ng/ ul
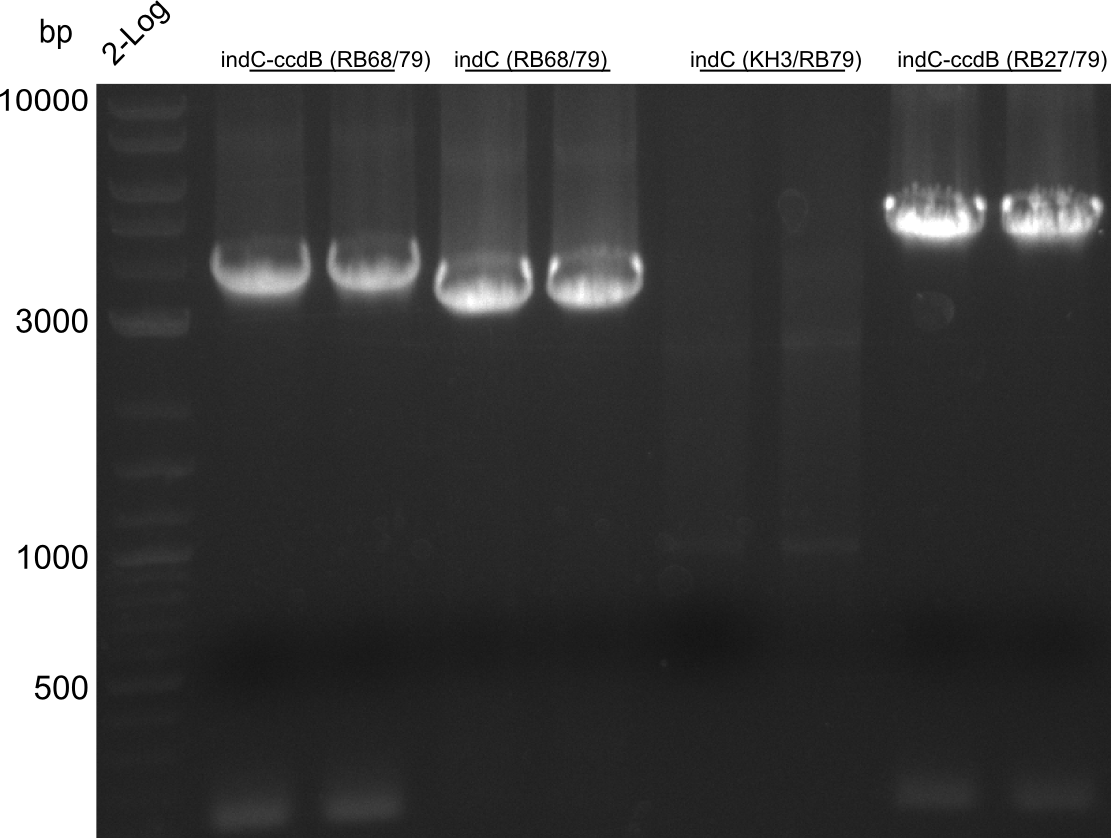
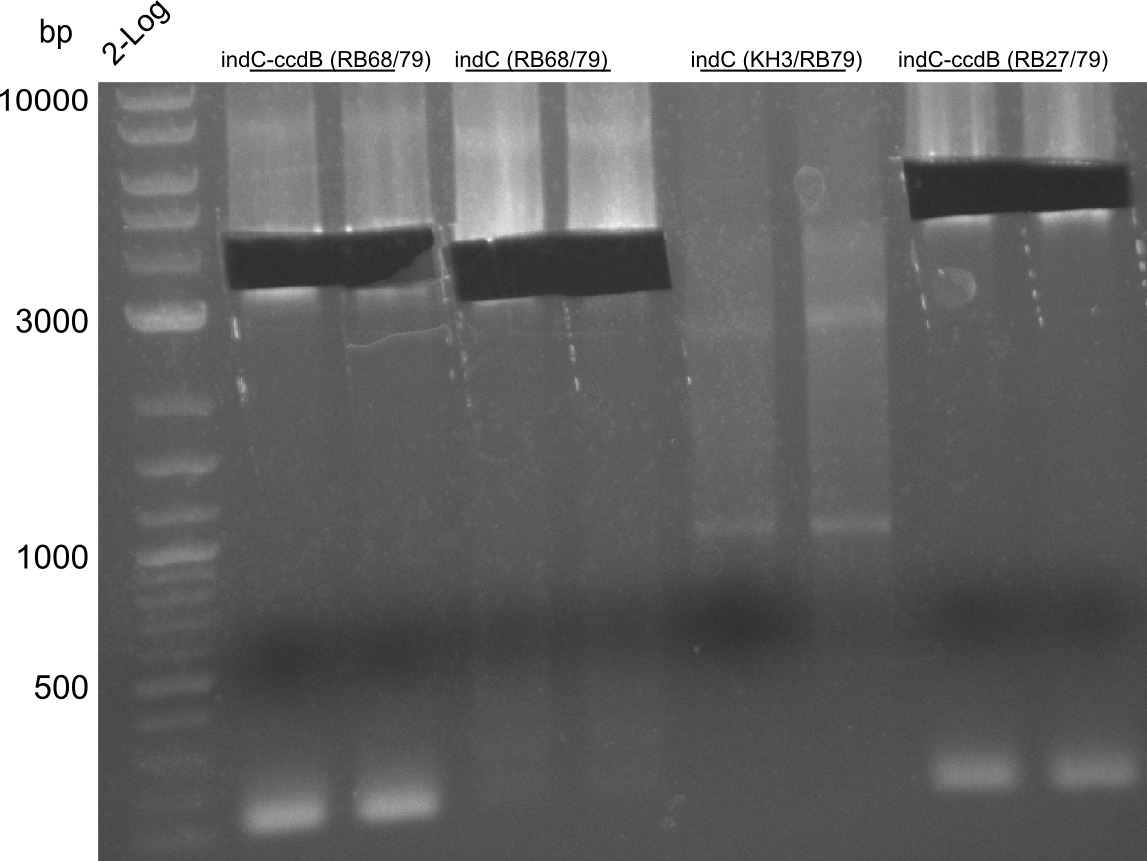
CPEC assemblies were performed for following plasmids with the respective fragments:
- pRB19: RB27/79 from pRB14 and RB21/22 from pSB1C3 midiprep
- pRB22: RB27/69 RB70/78 RB68/79 RB21/22
- pRB23: RB27/69 RB70/78 RB68/KH4 KH9/RB79 (pRB14) RB21/22
- pRB24: RB27/28 RB66/67 RB21/22
- pKH5: KH3/4 (pKH4) KH9/10
Transformed cells looked as follows:
Heidelberg 201309 CPECs
|
Colony screenings
Heidelberg 201309 CPECs screening
|
pRB23 assembly was unsuccessful, pRB22 is correct (plates turn blue), pRB24 seems to be correct (small deep blue colonies), pRB19 didn't work, pKH5 seems correct. We prepped the marked colonies. Sequencing showed that pRB22 is correct and doesn't carry endogenous RFC10 cutting sites any longer. Test digestion with EcoRI and SpeI also enforced this result.
We performed a PCR with Phusion HF and KH3/4 from pRB22, assembled it in a CPEC approach with ccdB KH9/10 and transformed OneShot cells.
Colony screenings and test-transformation in TOP10 cells showed that the assembly was successful and that we're able to shuffle the T-domains.
Minipreps
Minipreps of pRB15, pRB17, pRB18, pKH4 and pRB14 yielded:
- pRB15
- pRB17
- pRB18
- pRB14: 284.8 ng/ ul in 20 ul
- pKH4: 316.8 ng/ ul in 20 ul
5 ul of each pRB15, 17 and 18 were put on an agarose gel with small pockets.
Heidelberg 20130902 indC
|
PPTase plasmid minipreps are empty. We will try to assemble them again using a restriction strategy besides prepping them again. Repetitioin of the miniprep was successful and the plasmids were sent to sequencing.
Cotransformation
We did cotransformation of pKH4 with pRB15-18, respectively, and single transformation of pSB3K3, pKH4 and pRB15-18.
Restriction cloning for pRB15-19
| RB21/22 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| td 60 | 5 | |
| 72 | 50 | |
| 23 | 98 | 1 |
| 65 | 5 | |
| 72 | 40 | |
| 1 | 72 | 3.00 |
| 1 | 12 | - |
| RB27/46 in BioRAD T100 | ||
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| td 60 | 5 | |
| 72 | 70 | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 60 | |
| 1 | 72 | 2.00 |
| 1 | 12 | - |
Heidelberg 20130902 indC res
|
Heidelberg 20130902 indC res
|
svp was reamplified from short product
| RB21/22 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 65 | 5 | |
| 72 | 20 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
Heidelberg 20130902 svplong
|
Heidelberg 20130902 svplong
|
indC and pSB1C3 were digested with KpnI and NheI, sfp, svp, entD, delC and pSB3K3 with BamHI and NheI for two hours at 37 °C.
Heidelberg 20130903 digest
|
Heidelberg 20130903 digest cut
|
Unfortunately pSB3K3 contains an internal NheI cutting site. We repeated the amplification, purification, restriction digest and purification with BamHI and SpeI, which builds a scar with NheI.
Heidelberg 20130903 pSB3K3
|
Heidelberg 20130903 pSB3K3 cut
|
Ligation of pSB1C3 with indC(ccdB) was performed over night in a 10 ul reaction with 4 ul fragments each.
Transformation of ligation mixes were performed with 5 µl each in TOP10. Additionally 5 µl of ligation mix of pRB19 was transformed into OneShot cells.
Heidelberg 20130905 plates pRB15-18-ligation
|
Heidelberg 20130905 plates pRB19-ligation
|
Colony screenings showed following results:
We prepped pRB15, 17 and 18 and repeated the assembly of pRB16 from scratch. Therefore we used RB13/14 for amplification of svp from an S. verticilllus ATCC15003 liquid culture. Afterwards we reamplified this PCR gel extraction with RB29/30 and performed a restriction digest with BamHI and NheI to ligate the fragment with pSB3K3 (BamHI; SpeI).
Reevaluation of old MP pRB15-18, pRB21
Because we could observe small colonies with darkgray center on cotransformed pRB21 with all PPtases constructs (plates of 2013-08-27), we conducted a colony screen and a retransformation with 1 µl of previous MP of pRB15.2 (Konrad) and pRB16-18 (Ralf) and pRB21 (Ralf). Additionally a cotransformation with this old pRB21 MP with the old pRB15.2 as well with 2.5 µl of newly assembled pRB15 was performed.
Heidelberg 20130905 plates pRB15-18 pRB21
|
Heidelberg 20130905 plates CT pRB15-pRB21
|
The colony screening was performed using the primers VR and KH5. If the gray colonies carry two plasmids with indigoidine synthetase and activating PPtase separately an amplicon of size ~1.3 kbp. Otherwise if for some reason the PPtase and indigoidine synthetase are still on one plasmid, we could expect an amplicon of 2.1 kbp.
| KH5/VR in Biometra, 10 µl | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 95 | 120 |
| 30 | 95 | 30 |
| 60 | 30 | |
| 72 | 120 | |
| 1 | 72 | 240 |
| 1 | 12 | - |
The positive controls gave the expected bands. All picked clones also gave an expected band at 1.3 kbp which leads to the conclusion that they carry the indC-only plasmid pRB21. Because every third clone was a picked white colony we could also conclude, that not all transformants carry an intact PPtase plasmid.
T-domain exchange with different domain borders
We thought of a different way to set the T-Domain borders. Starting from the Pfam-predicted domain borders we made pairwise alignments to find amino acid sequences up- and downstream the predicted borders, that are identical in both the native and the brought in sequence. In the case of the exchange of indC and bpsA those new borders would be around 10 amino acids more on each side. We designed primers for this strategy.
Screenings
| Plasmid | Primer 1 | Primer 2 | Expected product [bp] | wrong product [bp] | additional test |
|---|---|---|---|---|---|
| pKH6 | VR | RB17 | 1070 | ||
| pKH7 | VR | RB13 | 1120 | ||
| pKH8 | VR | RB41 | 1000 | ||
| pKH9 | VR | RB64 | 1070 | ||
| pRB15_lig | VR | RB17 | 1070 | ||
| pRB16_lig | VR | RB13 | 1120 | ||
| pRB17_lig | VR | RB41 | 1000 | ||
| pRB18_lig | VR | RB64 | 1070 | ||
| pRB19_lig | VR | KH9 | 1900 | 2600 | |
| VR | RB17 | - | 1070 | ||
| pRB22_frag | digest | ||||
| pRB23_frag | VR | KH9 | 1900 | digest | |
| RB27 | KH4 | 2900 | |||
| pKH5 | VR | KH9 | 2100 | ||
| pRB24 | VR | KH5 | 2600 | ||
| VR | RB64 | 1070 | |||
| pRB7 | - | - | - | - | digest |
We repeated the screening of pKH8 with 20 colonies and amplified svp with RB29/30 from pKH7.
 "
"