Team:Greensboro-Austin/Smell degradation
From 2013.igem.org
Contents |
Introduction
p-Cresol (4-methyl phenol) is a volatile organic compound present in most mammal fæces, correlated with having an odor characteristic to humans as fecal and detectable at dilute concentrations (Zahn et al., 2001). The compound is present in its highest concentrations in pig manure and is formed in this context by the anaerobic degradation of the amino acid L-tyrosine, which occurs in the metabolism of the natural intestinal flora (Antoine, Taillieu, & Thonart, 1997; Mathus & Miller, 1995).
Pseudomonas putida
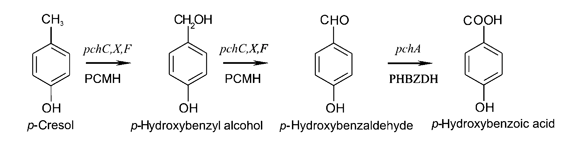
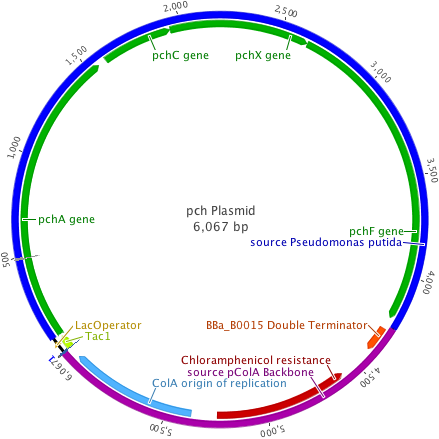
The pch gene cluster follows the protocatcechuate branch of the ortho pathway of p-cresol degradation, encoding for the enzymes p-cresol methylhydroxylase and p-hydroxybenzaldehyde dehydrogenase, which degrade p-cresol to p-hydroxybenzoic acid (Jõesaar et al., 2010). This pch cluster has been studied in multiple strains of Pseudomonas putida and other microorganisms. One of these P. putida strains, NCIMB 9866, contains the pchACXF gene cluster, its genes have been sequenced and annotated, and is readily available. If this gene cluster could be inserted into and expressed by a bacterial species that is as manipulable as Escherichia coli, it could become an effective tool for the bioremediation of p-cresol pollution.
Bacillus sp.
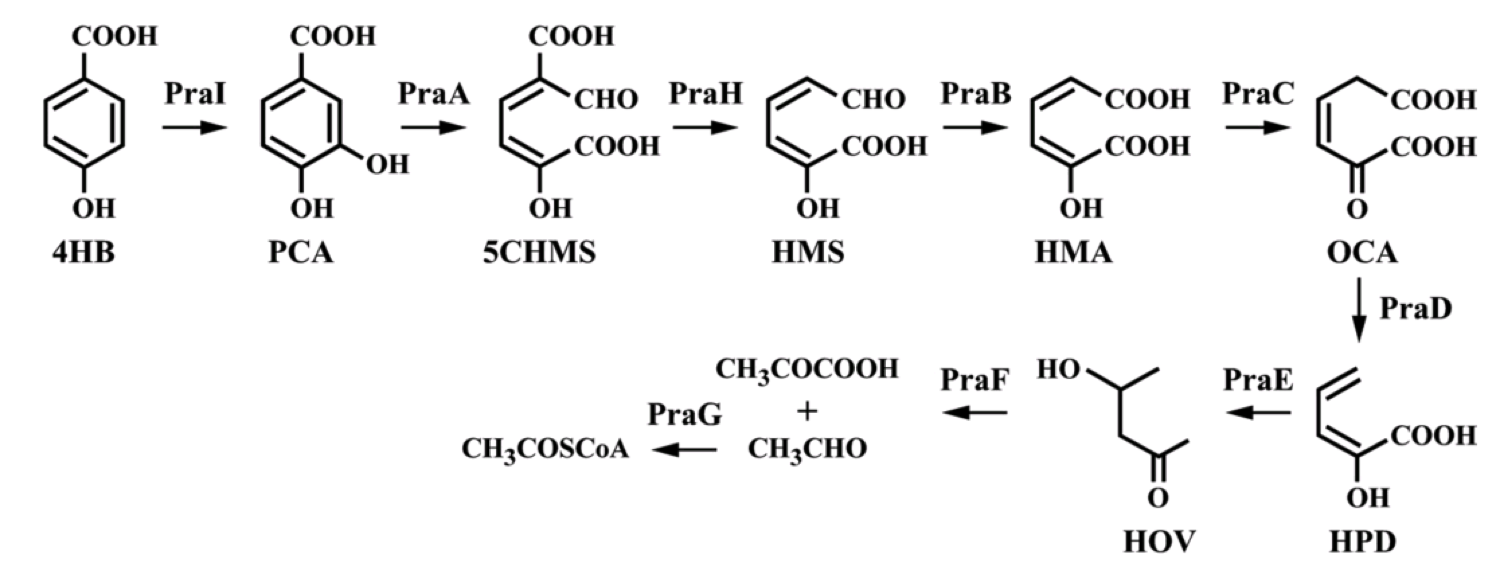
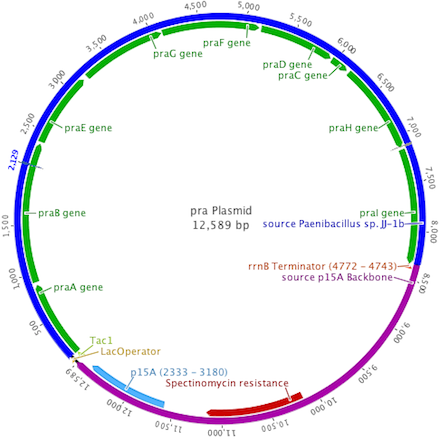
To create a self-sufficient bioremediation tool for p-cresol, the pch cluster will be accompanied by the pra cluster. This second gene cluster encodes for several enzymes that degrade the end product of the pch gene cluster, p-hydroxybenzoic acid, down a pathway that results in the formation of acetyl coenzyme A and pyruvate, which then feed into the citric acid cycle for energy production and cell growth (Crawford, 1975). The praABEGFDCHI gene cluster from the Bacillus sp. strain JJ-1b has been successfully expressed in E. coli, its genes sequenced and annotated, and is readily available (Kasai et al., 2009). A functional E. coli bacterium containing both the pch and pra gene clusters could be able to survive on p-cresol as its sole carbon source. It should be noted, however, that the pra plasmid is not necessary for deodorization of p-cresol. If successfully expressed, the pch plasmid alone would degrade p-cresol to the effectively odorless intermediate, p-hydroxybenzoic acid.
Clostridium difficile
p-Cresol has one final element of interest to humans: it is secreted by the dangerous microorganism Clostridium difficile when forming an infection in the intestine, which is common as a hospital acquired infection and can be fatal. The p-cresol secreted by C. difficile is toxic to almost all other microbes in the intestinal flora, severely disrupting gut function and allowing C. difficile to flourish in the absence of others (Dawson, Stabler, & Wren, 2008). An organism that could efficiently degrade the p-cresol being secreted might be able to restore gut function and inhibit the progression of the infection.
Strategy
Escherichia coli
The aforementioned gene clusters pchACXF and praABEGFDCHI will be cloned into separate plasmids and transformed into the single-gene knock-out strain of BW25113 E. coli that has had the gene coding for tryptophanase removed. Tryptophanase is an enzyme that converts tryptophan into indole, the compound responsible for the characteristic smell of E. coli. Without the ability to produce indole from tryptophan, the strain itself is effectively odorless.
Directed Evolution
In addition, the untransformed knock-out strain of BW25113 E. coli will undergo directed evolution to select for reduction of p-cresol toxicity.
References
- Kasai, D. et al. Uncovering the Protocatechuate 2,3-Cleavage Pathway Genes. Journal of Bacteriology 191, 6758–68 (2009).
 "
"