Team:DTU-Denmark/Notebook/14 August 2013
From 2013.igem.org
14 August 2013
Contents |
lab 208
Main purpose
- PCR for extraction of Nir2 from PAO1
- extract fragments for SPL and perform USER ligation
Who was in the lab
Gosia, Kristian, Julia, Henrike
Procedure
PCR for Nir2 extraction from PAO1
We tried to reproduce the successful extraction of Nir2 from PAO1. The extracted Nir2 fragment will be used as template for a screening PCR with USER primers.
primers: 42a and 42b
template: 1 colony of P. aeruginosa cells resuspended in 100 uL milliQ water
program: touchdown PCR from 64C to 57C(called U2 on Eppendorf)
- step 1 - 98C 10:00
- step 2 - 98C 0:20
- step 3 - 64C 1:00
- step 4 - go down 0.5C each cycle
- step 5 - 72C 3:00
- step 6 - GOTO step 2 for 14 cycles
- step 7 - 98C 0:20
- step 8 - 57C 1:00
- step 9 - 72C 3:00
- step 10 - GOTO step 7 for 20 cycles
- step 11 - 72C 5:00
- step 12 - 10C hold
The table shows the composition of each reaction:
| component (per reaction) | without additives | 5% DMSO | 2mM MgCl2 | 5% DMSO + 2mM MgCl2 | Negative control |
|---|---|---|---|---|---|
| dNTPs | 1uL | 1uL | 1uL | 1uL | 1uL |
| GC buffer | 10uL | 10uL | 10uL | 10uL | 10uL |
| Phusion polymerase | 0.5uL | 0.5uL | 0.5uL | 0.5uL | 0.5uL |
| MilliQ water | 31.5uL | 29uL | 29.5uL | 27uL | 32.5uL |
| template | 1uL | 1uL | 1uL | 1uL | - |
| FW primer | 3uL | 3uL | 3uL | 3uL | 3uL |
| RV primer | 3uL | 3uL | 3uL | 3uL | 3uL |
| DMSO | - | 2.5uL | - | 2.5uL | - |
| MgCl2 | - | - | 2uL | 2uL | - |
PCR for Nir2 with USER endings
We are still trying to amplify Nir2 with endings that are suitable for USER cloning with Nir1. The same reaction mix as yesterday was run on two new programs.
Sample 1-6: 63C annealing temperature
Sample 7-12: ramp from 65C-57C
If this PCR is not successful we suggest to try touchdown.
Gel purification
gel purify PCR products from constitutive promoter SPL, constitutive promoter ref and araBAD ref
User ligation and transformation for SPL project
Ligated and transformed the araBAD SPL, the araBAD reference promoter, the constitutive promoter SPL and constitutive reference promoter. For every fragment was performed the standard reaction, a reaction containing a clean-up step for small nucleotides before transformation and a negative control. Additionally a positive control was made with RFP from the BioBrick transformation efficiency kit.
Plated the whole reaction volume.
touchdown PCR for Nir2 with User endings
tried touchdown PCR for Nir2 with User endings with this program:
- step 1 - 98C 10:00
- step 2 - 98C 0:20
- step 3 - 72C 1:00
- step 4 - go down 0.5C each cycle
- step 5 - 72C 3:00
- step 6 - GOTO step 2 for 14 cycles
- step 7 - 98C 0:20
- step 8 - 65C 1:00
- step 9 - 72C 3:00
- step 10 - GOTO step 7 for 20 cycles
- step 11 - 72C 5:00
- step 12 - 10C hold
Results
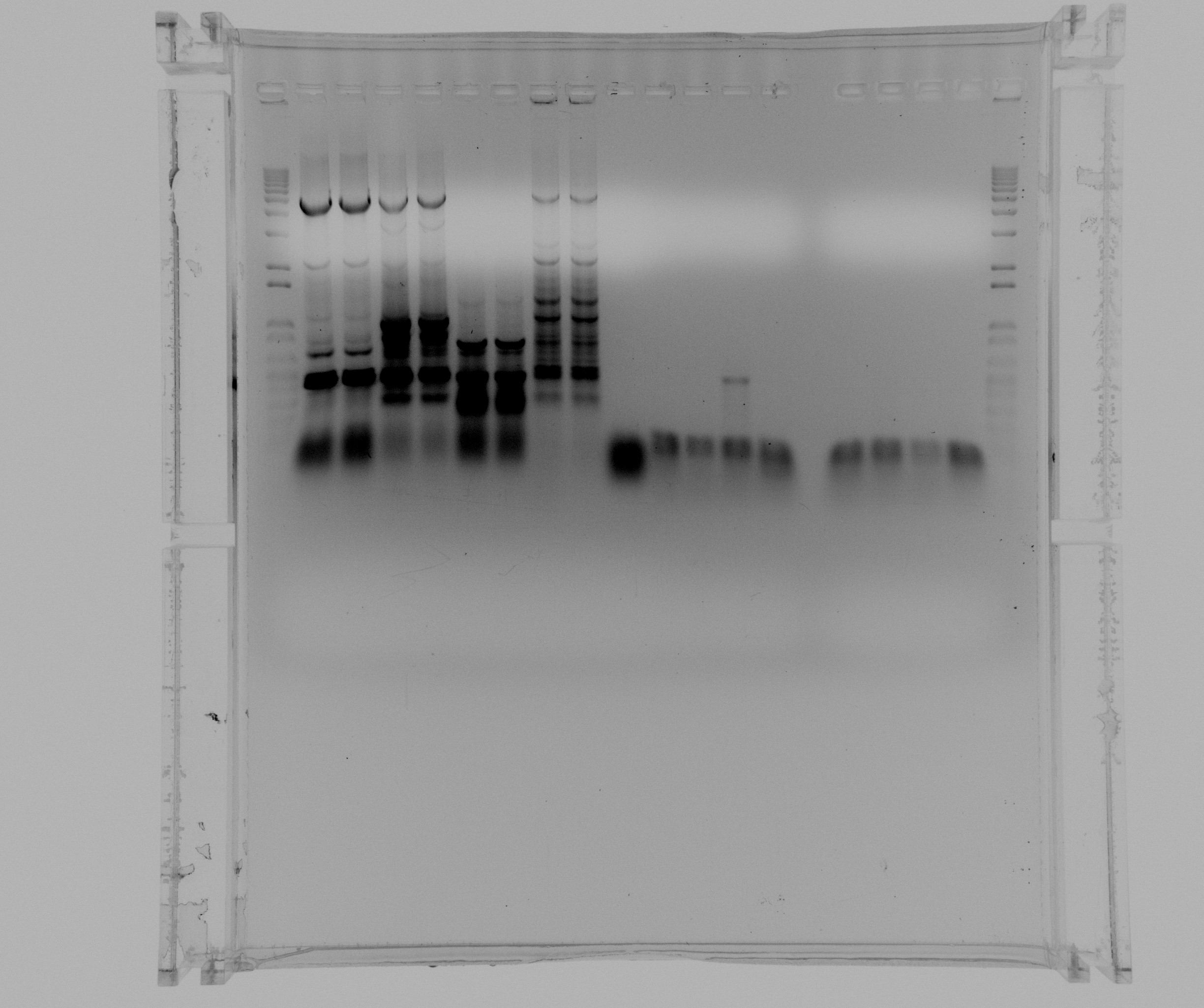
Purification gel
- 1 kb ladder
- constitutive promoter SPL
- constitutive promoter SPL
- constitutive promoter reference
- constitutive promoter reference
- araBAD promoter reference
- araBAD promoter reference
- 1 kb ladder
Gel on todays PCR
- 1kb ladder
- Nir2 extraction PCR without additives
- Nir2 extraction PCR without additives
- Nir2 extraction PCR with 5% DMSO
- Nir2 extraction PCR with 5% DMSO
- Nir2 extraction PCR with MgCl
- Nir2 extraction PCR with MgCl
- Nir2 extraction PCR with 5% DMSO and MgCl2
- Nir2 extraction PCR with 5% DMSO and MgCl2
- negative from Nir2 extraction PCR
- sample 1 Nir2 User PCR
- sample 2 Nir2 User PCR
- sample 3 Nir2 User PCR
- sample 4 Nir2 User PCR
- sample 5 Nir2 User PCR
- sample 6 Nir2 User PCR
- sample 7 Nir2 User PCR
- sample 8 Nir2 User PCR
- 1 kb ladder
Conclusion
We were able to amplify the Nir2 by extracting it from genomic DNA from P. aeruginosa and will purify tomorrow. The PCR of Nir2 with USER endings was not successful and will be repeated with a touchdown program before doing a gradient PCR.
lab 115
Main purpose
- Distinguish the proper amount of glucose substrate for DM minimal medium in order to grow the E.coli cells
Who was in the lab
Ariadni, Helen, Natalia
Procedure
Calculate different concentrations of DM Minimal medium with NH4Cl and glucose for overnight growing E.coli cells.
First we make one NH4Cl 0.916 M solution
- 2.45 gr of NH4Cl
- 50 ml of DM minimal medium
The table shows the composition of each tube before we add some cells:
| Concentration of glucose (%) | Glucose (gr) | NH4Cl (uL) | DM minimal medium (uL) | Total volume (mL) |
|---|---|---|---|---|
| 0.5 | 0 | 50 | 9950 | 10 |
| 1 | 0.0489 | 100 | 9900 | 10 |
| 5 | 0.4401 | 500 | 9500 | 10 |
| 10 | 0.9291 | 1000 | 9000 | 10 |
Results
Conclusion
Navigate to the Previous or the Next Entry
 "
"