Team:UChicago/Notebook
From 2013.igem.org
Contents |
June
Week 3
Tuesday, June 18, 2013
First iGEM Meeting of the Summer!
- All student members gathered in the lab.
- We toured the lab, prep room, and incubator room, and we discussed the lab rules.
- We also split into teams to discuss lab logistics: contacting funding sources, ordering reagents/materials, compiling protocols, recording our electronic lab notebook, and making plates/media.
Week 4
Wednesday, June 26, 2013
Planning Wet Lab Work
- We split into planning sub-teams to plan out different parts of the project with protocols, timelines, and ordering reagents.
- Plasmid Design
- Media and Competent Cells
- E. coli and B. subtilis Transformation
- Keratinase Assay and His tagging
- Running DNA gels, SDS-PAGE gels, and western blots
- We will also be holding weekly meetings with our graduate student advisers and biweekly meetings with our faculty advisers to provide updates.
- Wet lab work begins tomorrow!
Friday, June 28, 2013
Researching the Sequence of kerA
- Planning subteams continued to order reagents and compile protocols.
- The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT.
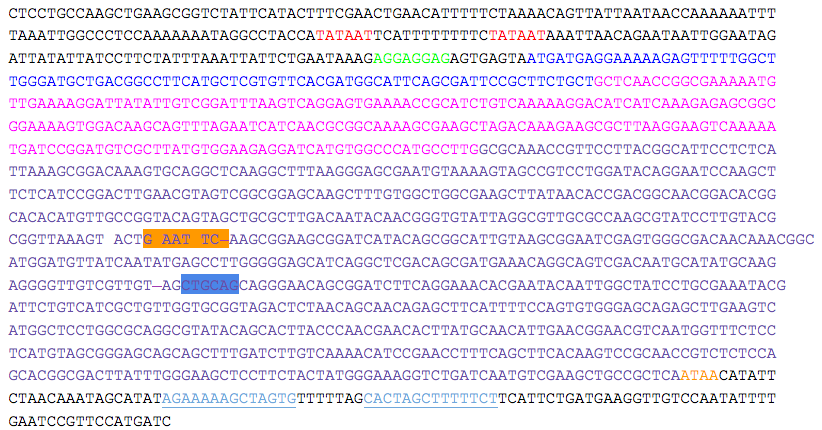
- kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995).
- Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated.
- EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick.
July
Week 1
Wednesday, July 3, 2013
BioBrick Design Presentation and Grad Adviser Meeting
- Designed a biobrick with:
- C terminus His-tagged kerA
- CATCATCATCATCATCAT
- Prefix: gaattcgcggccgcttctagag
- Suffix: tactagtagcggccgctgcag
- Moreover, the bio brick must include
- Eliminated EcoRI and PstI restriction sites from kerA nucleotide seq
- EcoRI (gaattc--814) and PstI (ctgcag--978)
- Conserved aa seq + biobrick prefix + suffix
- Secretion signal peptide
- Inducible promoter
- We also presented a feasible timeline of our project, which included:
- Assembly of biobrick and transformation into E. coli
- Transformation of B. subtilis
- Keratinase isolation and keratinase assays
- kerA mutagenesis for optimization of the keratinase
- We also split up basic lab duties!
- Ivan & Ethan are in charge of the IBC protocol.
- We can’t start wet lab work until our IBC protocol is finished!
- Ethan is in charge of minutes.
- Susan is in charge of ordering invoices.
- Annie is in charge of emailing advisors and members to plan meetings
- Chloe is getting the B. subtilis gene that expressing keratinase.
Thursday, July 4, 2013
Milk Agar Plates
- The purpose of milk agar plates is often to detect protease activity, including the activity of keratinases.
- A measurement of any baseline protease activity of our parent B. subtilis strain (WB700), which does not endogenously express any keratinases, is desired.
- Excess protease activity of our parent B. subtilis strain may prevent our use of milk agar plates as an assay for keratinase expression of our future transformants with the kerA gene.
- Therefore, the parent B. subitilis strain (WB700) was streaked on the milk agar plates to observe baseline protease activity that could prevent us from detecting keratinase expression after transformation.
- Milk Agar Plates (in LB):
- Ideally, there should be a minimal zone of clearing around our B. subtilis colonies.
- Next Steps:
- Observe the results of our milk agar plates with B. subtilis colonies
Friday, July 5, 2013
Primer Design for kerA biobrick
- Designed primers for directed mutagenesis of our kerA biobrick using [http://www.bioinformatics.org/primerx/ PrimerX]:
- Primer pair 1 (EcoRI mutation, aat --> aac, asparagine)
- a. Forward: 5' GGTTAAAGTACTGAACTCAAGCGGAAGCGG 3'
- b. Reverse: 5' CCGCTTCCGCTTGAGTTCAGTACTTTAACC 3'
- Primer pair 2 (Pst1 mutation, gca-->gcg, alanine)
- a. Forward: 5' GTTGTCGTTGTAGCTGCGGCAGGGAACAGCGGATC 3'
- b. Reverse: 5' GATCCGCTGTTCCCTGCCGCAGCTACAACGACAAC 3'
- Used APE for checking codons and mutating restriction sites
Saturday, July 6, 2013
Primer Design Part II
- Designed primers to isolate non-coding and coding sequence of kerA and add prefix and suffix
- forward primer + xbaI site:
- 5' TATCTAGAGGTCTATTCATACTTTCGAA 3'
- reverse primer + speI site (reverse complement):
- 5' ATTGATCATTGGACAACCTTCATCAG 3'
- We still need to design additional primers that will include the secretion signal peptide (in case it needs to be secreted out of B. subtilis) and kerA (From start to stop codon).
- These primers should include igem biobrick prefix and his tag (before stop codon) + suffix.
Week 2
Monday, July 8, 2013
Milk Agar Plate Results
- The results of our milk agar assay from 7/04/13 of the non-transformed B. subtilis strain WB700:
- The zone of clearing around the B. subtilis colonies indicates that our parent strain breaks down milk. Its prominent endogenous protease activity prevents us from using the milk agar assay as a screen for successfully transformed kerA-expressing B. subtilis.
- Alternative method to screen for transformed B. subtilis needed:
- The feather assay used in Saha and Dhanasekaran’s 2010 paper, “Isolation and Screening of Keratinolytic Actinobacteria form Keratin Waste Dumped Soil in Tiruchirappalli and Nammakkal, Tamil Nadu, India”
- Quantitative assay using azokeratin
Our future plans:
- For our promoter for kerA, Karyl, one of our graduate advisors, recommended P43, a constitutive promoter, to transform into B. subtilis.
- We also decided to use pUB110, a high copy number plasmid used in S. aureus and B. subtilis, to introduce kerA into our parent B. subtilis strain.
- pUB110 will also be made into a biobrick.
- We have to choose a suitable plasmid (either pSB1A3, pSB1C3, pSB1T3, or pSB1K3.m1) for our E. coli transformations.
Wednesday, July 10, 2013
Grad Advisor Meeting on further developed Keratinase Project
- We presented the primers we designed to our advisers, who suggested we double check them for self dimerization and 3’ overlaps with:
- http://www.idtdna.com/analyzer/applications/oligoanalyzer/
- http://bioinfo.ut.ee/primer3/
- We discovered that our forward primer forms a self-dimer and must be redesigned.
- We also need lysozyme to digest the peptidoglycan of B. subtilis in order to miniprep it.
- We decided to use the iGEM plasmid pSB1K3.m1 for our E. coli transformations.
- To screen for keratinase activity, instead of using the feather assay, which takes up to a month for visible results to appear, we decided to use the azokeratin assay as the secondary screening method.
Thursday, July 11, 2013
Meeting with Purdue iGEM team
Get Excited for the UChicago X Purdue collaboration! Our first meeting with the Purdue team went well:
- The Purdue team will send us a form to provide feedback on their igem biobrick characterization and protocol forms on Monday.
- The biobrick forms are in PDF format.
- Their biobrick characterization contains info like biobrick sequence, plasmid and microorganism in which it’s inserted, ect.
- The protocol form is a checklist of the protocols carried out to characterize the biobrick. It doesn’t contain much info.
- Nora asked them to add a section stating the purpose for using a specific protocol, as in, what specifically about the biobrick are they trying to characterize.
Week 3
Week 4
Tuesday, July 23, 2013
Today we made LB media with kanamycin and chloramphenicol. There are 7 plates of each kind of antibiotic marker left over; we put them in the 4ºC fridge. We autoclaved all the media that we made today and yesterday.
We transformed the plasmid with promoter Veg (in the pSB1C3 vector) into TOP10 E. coli competent cells. We got the DNA (part BBa_K143012) from well 23B on the distribution kit. The bacteria have been plated onto a chloramphenicol plate. They have been left to incubate at 37ºC in Nora’s lab (GCIS W519).
Future plans:
- Select a few colonies and inoculate overnight in 3 mL cultures (These will eventually be used to make glycerol stocks and for DNA miniprep). Do this later on in the day... ~3 pm onwards.
- Store the plate in 4ºC afterwards in case it is needed in the future.
- Make liquid LB media (no agar!) with 50 µg/mL chloramphenicol.
Wednesday, July 24, 2013
2 colonies were selected from yesterday's pVeg transformants to be grown in an overnight liquid culture. However, we redid the transformation using a slightly different procedure because there were very few transformants:
- Let the cells thaw on ice.
- Add your plasmid to the competent cells.
- Leave on ice for 10 mins.
- Place on the 42ºC heat block for 45 s to heat shock.
- Put on ice for 2 minutes.
- Add 1 mL SOC media.
- Incubate in the 37ºC shaker for 1 hour.
- Centrifuge at 8 rpm for 30 seconds. Decant all but ~100 µL supernatant and resuspend.
- Plate onto chloramphenicol (or antibiotic of choice) plates and use glass beads to spread out the colonies.
Future plans:
- Make DH5a competent cells; our poor transformation results could be due to incompetent cells.
- Isolate Pveg (for B. subtilis) promoter from transformants grown overnight
- Do DNA miniprep of the transformants
- Cut w/ EcoRI and SpeI
- Run in 0.9% or 1% agarose gel (make gel w/ large wells, load 50 uL of sample)
- If there is a band of right size of Pveg, do a gel extraction
- Put at -20ºC
- If there is no correct band, set overnight cultures of transformants from the transfromation done by Ivan today (7/24/13)
- And for Friday's meeting (7/26/13)
- Isolate Pveg (for B. subtilis) promoter as described above if it was unsuccessful the day before. Use the overnight cultures set up on 7/25/13.
- Check if the transformation w/ the E. coli constitutive promoter was successful. If yes, store plate at 4ºC.
Friday, July 26, 2013
To do:</i>
- Make overnight cultures of colonies from Annie’s transformation.
- Transform 18O and 18E again, this time into the competent cells made yesterday by Ethan and Annie. Those plasmids should be in the -20ºC.--DO MONDAY, Ethan’s doing with transformation efficiency 7/29
- Check the efficiency of our competent cells by using the transformation efficiency kit from iGEM (check the instructions on the iGEM wiki).--DO MONDAY
- Do a gel purification of the digestion of the PVeg promoter & check the size.
- The digested DNA is sitting in the fridge. Use ALL of it.
Week 5
Monday, July 29, 2013
<i>To do:
- Set O.N. culture of 18O (2X 3mL)--done by Annie (18O is the E. coli constitutive promoter)
- Check transformation efficiency, 18E of competent cells made by Ethan
- Run gel with Pveg digestion (cut w/ EcoRI and SpeI)
- Redo the digestion with 10uL of DNA miniprep of Pveg w/ EcoRI and SpeI for 2 hr
- Run in 2% agarose
- If there are bands--purify promoter
- If there are no bands, set up O.N. culture of Ivan’s transformation of Pveg
- Set O.N. culture of B. subtilis BD 366 w/ pUB110 (2X 3mL)--done by Nora
Getting done today:
- Re-digest pVeg, digest 5ul of miniprep 1 2hrs at 37 deg C:
- 2 ul 10X buffer 2.1
- 1 ul EcoRI HF
- 1 ul SpeI
- 5 ul mini prep 1
- H2O to 20 ul
- Also set previously done digest by Klevin for 2 hrs at 37 deg C
--Done by Annie and Nora
- Transformed 50 uls of new competent cells and old competent cells with 0.5 ul of 18E resuspension and plated 50 ul of new competent cells as a negative control
--Done by Ethan
Next Steps:
- Run digest of Pveg on 2% agarose gel
- Do miniprep of B. subtilis pUB110
- Do miniprep of 18O transformants
Tuesday, July 30, 2013
- Ran gel of Pveg miniprep digest (Klevin’s at total 3hrs, and Nora’s/Annie’s at 2hrs digestion) with EcoRI and SpeI
- 10 ul ladder
- 20 ul of samples + 5 ul of Orange G dye
- Final gel: Nora’s and Annie’s (N&A) had faint band at 100 band marker, we assume this is the fragment
- Nora purified gel fragment, with Qiagen kit, eluted in 30ul water in order to obtain a higher concentration of DNA.
- Miniprep were prepared of 2 B. subtilis samples, pUB110 and 18O. They were stored at -20ºC.
- A transformation of 18E will need to be redone; incorrect selection marker was used.
- Ethan is testing the competency of the TOP10 cells he made.
Wednesday, July 31, 2013
gBlock Fragment Gibson Assembly calculations:
| total fmol (resuspended in 20 ul H2O) | final concentration | |
| gene block 1 | 648 fmol | .032 pmol/ul |
| gene block 2 | 455.4 fmol | .023 pmol/ul |
Gibson assembly mix (Trial 1):
- 4 ul gblock 1
- 6 ul gblock 2
- 10 ul master mix
We suspended pub110 insert oligonucleotides in H2O (to 100 uM) and annealed them at 94°C to make duplex dna
Next steps:
- Check the concentration on the oligos against the duplex DNA on the nanodrop to confirm annealing.
August
Week 1
Thursday, August 1, 2013
- Nora’s running PCR for Pveg, 18O, kerA assembly in Kron Lab
Concentration results from nanodrop:
- Forward pUB110 ssDNA: 1596.12 ng/µL
- Reverse pUB110 ssDNA: 1913.96 ng/µL
- Annealed pUB110 dsDNA: 2114.50 ng/µL*
| sample | ng/ul | A260 | A280 | A230 | 260/230 | extinction coefficient |
| oligo-f | 1596 | 48.4 | 25.6 | 22.3 | 2.17 | 33 |
| oligo-r | 1914 | 58.0 | 30.0 | 26.4 | 2.2 | 33 |
| annealed | 2159 | 42.3 | 22.3 | 13.8 | 2.25 | 50 |
A = E * b * c
C_annealed = [A / (50 * b)] 2159*(50/33) = 3271
Mass concentration of forward and reverse oligo mixture (ng/ul) = (1596 + 1914)/2 = 1755 ng/ul
- If none of the DNA anneals, the extinction coefficient is 33 and the nanodrop should read 1755*(50/33) = 2659 ng/ul
- If DNA was completely annealed, the extinction coefficient is 50 and the reading on the nanodrop should be 1755 ng/ul
- Therefore, the percentage of annealing is (2659 - 2159)/(2659 - 1755) = 500/905 = 55 %
The assembled DNA was then digested in the following conditions:
- 0.5 ul pUB110 insert
- 0.5 ul NdeI
- 0.5 ul AflII (N)
- 1.5 ul 2.1 buffer
- 12 ul H2O
- 5 ul pUB110 miniprep #1
- 0.5 ul NdeI
- 0.5 ul AflII (N)
- 1.5 ul 2.1 buffer
- 7.5 ul H2O
Friday, August 2, 2013
- Today we set up digests of pUB110 and 18O.
- Conditions:
- 18O promoter:
- 5 ul 18O miniprep insert
- 0.5 ul EcoRI
- 0.5 ul PstI
- 1.5 ul 2.1 buffer
- 7.5 ul H2O
- Incubate at 37°C beginning at 3 pm
- pUB110 promoter:
- 5 ul pUB110 miniprep insert
- 0.5 ul Afl II (blue-labelled)
- 0.5 ul Nde I (Nora’s)
- 1.5 ul 2.1 buffer
- 7.5 ul H2O
- Incubate at 37°C for 2 hrs
- Yields of pVeg fragment DNA have been low; we will start over by picking more pVeg transformant colonies and cloning more DNA.
Saturday, August 3, 2013
Sunday, August 4, 2013
Week 2
Monday, August 5, 2013
- Digest pUB110 promoter for Pre gene (bit.ly/16v7PRO a planning doc):
- One of the enzymes may have lost activity, so use NdeI sample taken from Nora instead
- 7.5 ul H2O, nuclease-free
- 5 ul pUB110 miniprep insert, mp1
- 1.5 ul 2.1 buffer, labelled “in-use”
- 0.5 ul Afl II, blue-labelled Ethan’s
- 0.5 ul Nde I, Nora’s
- incubate at 37°C at 1pm to 3pm, placed in 4°C
Ran a gel of the following DNA samples:
- lane 1: pVeg mp1 (2 ul)
- lane 2: pVeg mp2 (2 ul)
- lane 3: pVeg mp3 (2 ul)
- lane 4: pVeg mp4 (2 ul)
- lane 5: empty
- lane 6: 18O digest (15 ul)
- lane 7: ladder (8 ul)
- lane 8: pUB119 digest (15 ul)
Based on the results, it seems that none of our digests are working. It may be that the enzymes we are using have lost activity, so we may have to obtain fresh ones.
The following transformations were carried out by Annie:
- BBa_K314103 → Plate 1 : 4D in pSB1C3 (1638 bp)
- BBa_K314100 → Kit 1 : 1G (518 bp)
- BBa_K541501 → 11E
- Plated 500ul per plate, incubated o/n in Nora’s lab
- Shaker was off when we arrived so unsure if that will affect anything...
In other news, the BL21 competent cells were dropped off by Karyl.
To Do:
- Run gel of sample from gel purified pUB110 from last week
- Re-do Digestion of pUB110 and single-enzyme digestion of pUB110, and digestion of pVeg miniprep (overnight) (Ethan)
- Gel purification of last week’s questionable pUB110 fragment (Klevin)
Tuesday, August 6, 2013
- Running the gel with inserts
- Running 2% agarose gel at 120 volts
- Lane 1: biobrick insert
- Lane 2: puB110 from gel extract (Klevin’s)
- Lane 3: pub110 from gel extract (Ethan’s)
- Lane 4: skipped
- Lane 5: pub110 insert forward
- Lane 6: ladder
All but pUB110 insert forward ran off of the gel (sorry!) Rerunning:
- Lane 1: ladder
- Lane 2: KerA biobrick
- Lane 3: pub110 insert forward
- Lane 4: pUB110 from gel extract (Klevin’s)
- Lane 5: pUB110 from gel extract (Ethan’s)
- Klevin set up the overnight culture of the casettes
- Susan gel extracts the PB110
- Ethan will pick up reagents for B. subtilis transformation
Advisor Meeting
- Find distribution part with only pVeg and RBS no S.S.
- Figure out how to buy keratin (non-hydrolyzed)
- Figure out how to make keratin infused plate
- Figure out what the f1 origin does, does it allow high copy number?
- Figure out if the Pre gene is necessary by trying B. subtilis transformation with pUB110 undigested
- Double check if pVeg (with the S.S.) is in frame with KerA gene; try these combos, all with pVeg + RBS
- KerA + pVeg, both with S.S.
- KerA + pVeg, with pVeg only with S.S.
- KerA + pVeg, with KerA only with S.S.
- Feathers dissolve in DMSO and precipitate it in acetone (Source: http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2672.1999.00485.x/full)
- Characterization of alkaline protease from Bacillus: http://link.springer.com/content/pdf/10.1007%2FBF00172544.pdf
- Japanese 1989 assay for Keratin: http://link.springer.com/content/pdf/10.1007%2FBF00263997.pdf
- Making keratin agarose plates 1985: http://www.sciencedirect.com/science/article/pii/0003269785902817
- Making keratin powder (first step of making keratin sponges): http://www.sciencedirect.com/science/article/pii/S0142961203010846
- Add picture of 11E in slide?
- linker insert (prefix/suffix), we need to miniprep it and check it on gel to see if it’s there, put in excess. If that doesn’t work, phosphorylate or titrate it
- NEED TO DO B. SUBTILIS TRANSFORMATION ASAP WITH PUB110
- Will B. subtilis secrete KerA as the same as licheniformis
- Figure out another assay for keratinase secretion
- replica plate to filter paper
- ust be unhydrolyzed
- google “high yield purificwool assay?
- recipes for embedding keratin
- grind up protein to put in agar to see clearing zone
- check for cheapest source, keratin from ‘body building’ powder
- feather powder
- keratin mation of keratin from wool fiber” req. formic acid
- random vs. non-random mutagenesis
- Screening Fungi for synthesis of keratin expre., feather meal http://www.vsrdjournals.com/vsrd/Issue/2011_3_Mar/2_Tom_Sinoy_Research_Article_Mar_2011.pdf
- Wool to keratin
- Find papers where they use them for sole carbon sources
Wednesday, August 7, 2013
- Competent cells have been made!
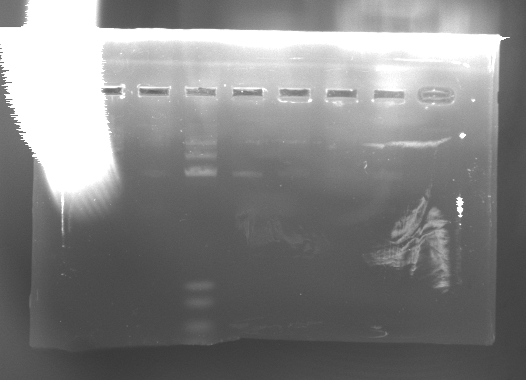
- Ran 1% gel with 6 minipreps of 11E, 1G, and 4D. The results are good. Image is in Google drive.
- lane1: ladder (8ul)
- lane2: 11E mp1 (2ul)
- lane3: 11E mp2 (2ul)
- lane4: 1G mp1 (2ul)
- lane5: 1G mp2 (2ul)
- lane6: 4D mp2 (2ul)
- lane7: 4D mp1 (2ul)
- Set up pUB110, pUB110 linker digestion o/n → see 8/8 for photo of digest- Nora
- Set up B. subtilis o/n culture- Ethan
- Ethan heat shocked Ivan’s competency cells 37ºC for 40s and then heat shocked again at 42ºC for 40s. In between heat shocks, he put it on ice. Transformed competency kit as test.-Ethan
- Requested Pveg + RBS part from iGEM, will arrive Tues 8/13 in agar slab, just plate and culture-Alice
Nanodrop concentrations
- Linker gel extract: 115.2 ng/ul (definitely should be visible on gel)
- pUB110 gel extract, more concentrated: 4.5 ng/ul (too low)
-Alice
Notes from meeting with Karyl
- Karyl suggests using quikchange to add his tag and suffix (no linker necessary)
- Need unmethylated and maybe use DpnI to eliminate template plasmid DNA
- Depending where active site is in KerA, quikchange may not work (ie if the active site is near the ends, there might be mutagenesis of the active end)
- If we have time, there are other peptides to consider besides the His-tag
- Perhaps try 3% gels for promoters
- Karyl says that our B. subtilis transformation protocol is okay to use
Thursday, August 8, 2013
- Digesting 2.5 and 5.0 ul of pub110 (miniprep # 2) starting at 12:45 pm--will leave over the weekend.
- If eluted with water, can be used for PCR.
Friday, August 9, 2013
pUB110 digest (3rd Trial)
- Digested 2.5 and 5.0 ul of pUB110 (miniprep # 2) starting at 12:45 pm.
- Digestion was left in incubator over the weekend
- Note: for better PCR results, elute with water
Week 3
Monday, August 12, 2013
Bacillus subtilis WB700 Transformation:
- Grew up overnight culture of B. subtilis strain WB700 in LB (to OD600 = 5.3) for transformation
- Made two 25 ml tubes of pre-transformation medium and added subtilis cells 1 and 2 ml respectively to OD .25 and .50
- note: The OD 0.5 tube fell off of the shaker some time in the first 3 hours of incubation and fell behind the 0.25 OD tube.
| Time (h) | OD 600 (sample 1) | OD 600 (sample 1) |
| 0 | 0.25 | 0.5 |
| 4 | 1.78 | 0.76 |
| 6 | 2.7 | 1.9 |
- After the 6 hours we spun the cells down in two 25 ml tubes and resuspended in 5 ml of the supernatant + 1 ml (sterile) glycerol
- Took 0.2 ml of resuspended cells and add to 1 ml (per transformation) of transformation medium
- Added DNA for transformation to tubes
Transformation:
| Cell Sample | ul of 100 ng/ul pUB110 miniprep |
| 1 | 0 ul (- control) |
| 1 | 1 ul |
| 1 | 5 ul |
| 2 | 1 ul |
| 2 | 5 ul |
- Then, we
- Froze the rest of the competent cells in liquid nitrogen in case the transformations work
- Incubated transformations for 2 hours at 37°C
- Spun down the transformation mixture, resuspended in 50-100 ul, and spreaded on kanamycin plates
Overnight digestions of kerA:
- 10 ul kerA
- 1.5 ul digest buffer
- 0.5 ul EcoRI (F.D.)
- 0.5 ul SpeI (F.D.)
- 2.5 ul H2O
Overnight digestions of pUB110
- 7.5 ul pUB110
- 1.5 ul copycutter 2.1 buffer
- 0.5 ul AflII (from nora)
- 0.5 ul Nde I (F.D.)
- 5 ul H2O
Tuesday, August 13, 2013
- Running out of pUB110 DNA, so we inoculated some overnights to clone some more.
Next steps:
- Miniprep of 3 tubes of pUB110 tomorrow.
- O/N culture of pSB1 C, and K plasmid backbones for E. coli
Wednesday, August 14, 2013
To Do:
- Transform pSB1A3 into competent DH5a cells
- Miniprep pUB110 from B. subtilis O.N. culture, then digest with AflII and NdeI to get rid of the Pre gene.
- Make more kanR agar plates because Ethan has taken the rest for B. subtilis transformation
- Make for LB broth and sterile-ly aliquot to 15ml tubes, to prevent any future large contamination issues.
- Need to make spreadsheet to log the progress of each BioBrick step
- Figure out how to submit DNA samples for sequencing.
- Order primers after Karyl has checked them.
- Advisor meeting today 7pm, some questions we'd like to address:
- Our his-tag is all CAT, will that be an issue?
- What B. subtilis strain can we borrow from Karyl that contains no knocked-out proteases?
- Team organization--how to split up work efficiently.
- Updates on putting kerA in Amp plasmid backbone and linker and pUB110 together.
Ask the listhost questions if you have them and when you have answers, reply all! Comment in the lab notebook with questions and reply if you have answers!
- We did minipreps to isolate more pUB110 DNA from the O/N cultures and set up a digestion. Enzymes should be heat inactivated at 80C prior to ligation.
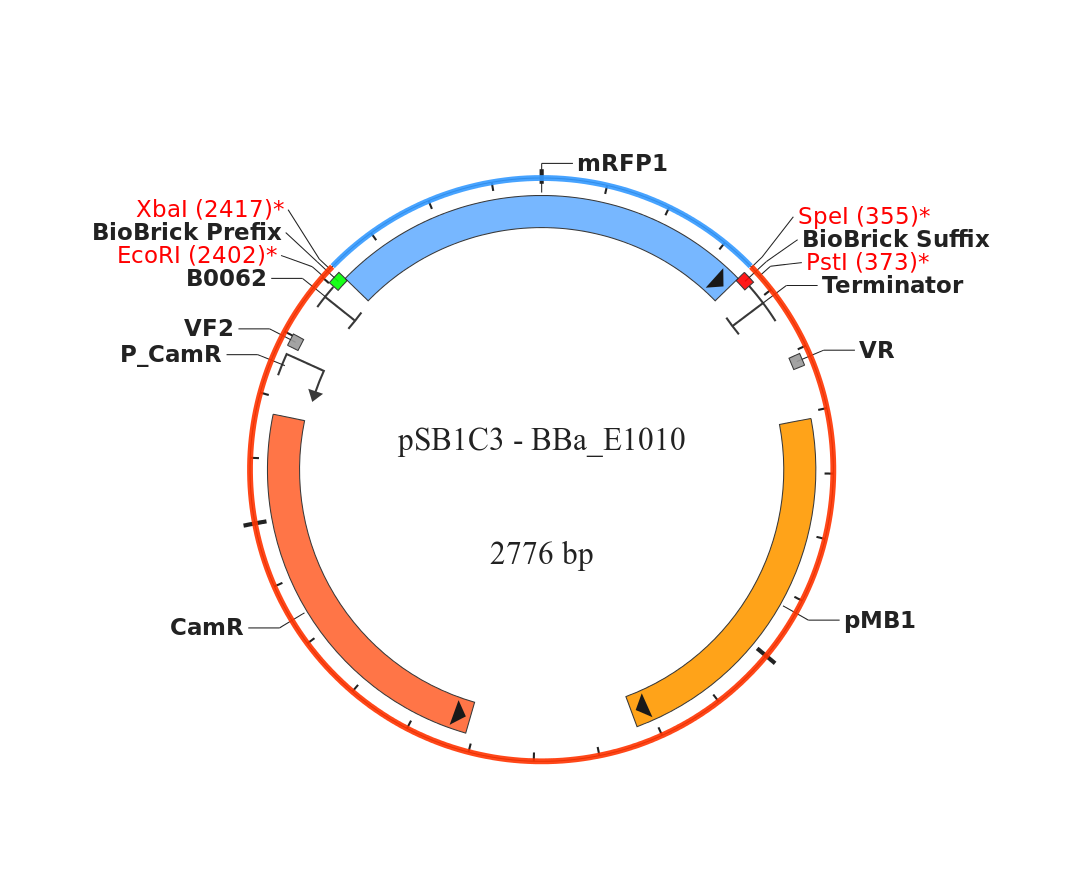
- We Found a Biobrick for RFP to put into B. subtilis, which we are doing to check for expression in B. subtilis. We’re using BBa_E1010 in pSB1C3, which has chloramphenicol resistance. We chose the C resistant one over the kanamycin resistant plasmid because we have more plates that are C resistant. The biobrick is located: plate 3, well 6G-Nicole.
Thursday, August 15, 2013
Digest of pUB110 and kerA-pSB1A3 Ligation
B.subtilis Backbone
- Ran a 1% agarose gel of 8/14’s pUB110 miniprep #2 O.N. digest, started 12pm (for eventual gel purfication)
- Used GelPilot loading dye, 5x from QIAGEN kit, 120V 12:47pm -
- Loading 7.5ul loading dye to 30ul of pUB110 digest
- lane 1: ladder (8ul)
- lane 2: pUB110 digest + dye (~19ul)
- lane 3: pUB110 digest + dye (~19ul)
- Results of the gel reveal that the digest was not successful. There was only one band around 4.5kb region, meaning no digestion occurred.
- Re-digested 8/14 pUB110 mp2 and mp1
| Item | Vol (ul) |
| 2.1 buffer | 3 |
| H20 | 15 |
| AflII Enzyme | 1 |
| NdeI Enzyme | 1 |
| pUB110 | 10 |
- Incubated in 37C overnight
- Note: Do not take the enzymes out of the freezer until you need them. Add them near the freezer. If this second digest doesn’t work, then there may be problems with the enzymes.
- We need more AFLII enzyme.
Restocking
- Made 100ml stock EtBr bath and used empty tip top to hold the bath. Remember to cover in aluminum foil. 5ul EtBr/100ml ddH2O.
- Since our concentrations are so low, we need to alter our gel purification protocol:
:*Remember to heat up elution buffer to 50C before use and let the binding buffer sit on the column for a few minutes, let the EtOH evaporate off too.
- Try using smaller columns and eluting twice with warm elution buffer. Take the total volume you want and divide it in half. First round of elution, let the EB sit for 10min and then spin down. And then do it again. The problem with the spin column 50 tubes is that the silica is too large compared to the volume of elution going through, so DNA tends to get struck. Flow-through is probably not the problem.
- Note: Gel purification does not work with 2% agarose gels
- Note ON AGAROSE GELS: It’s okay to reuse agarose gels, we just need to run the sample off. They can be re-used for a few weeks. The EtBr will need to be added to the TAE buffer in the apparatus though and re-added each time.
E.coli kerA Plasmid.
- Ligated kerA to pSB1A3, started 12pm, and transformed into E. coli
- First, Heat inactivated SpeI (from 8/14’s digestion) at 80C for 20min: 12:10pm - 12:30pm
| Ligase buffer (fixed) | 1ul |
| T4 DNA ligase (fixed) | 0.5ul |
| Vector (pSB1A3) 2155bp, 25ng/ul | 4.7ul |
| Insert (kerA) 1182bp, ~50ng/ul (assumption) | 3.8ul |
| Total | 10 ul |
- Incubated at RT for 20min, took 2ul for transformation of DH5a cells.
- Incubated with competent cells 4:10pm - 5:10pm and then plated on ampR plates.
- The rest of the ligation mix is incubating for 3:30pm-6:05pm and then heat to inactivate ligase, 65C for 10min. This ligation mix can be used again if the prior transformation doesn’t work.
E.coli Cassettes, B.subtilis Promoter.
Digest from Aug 8 photo taken from 8/8 lab notebok
lane1: ladder (8ul); lane2: 11E (2ul); lane3: 1G Cons cassette (2ul); lane4: 4D Lac cassette (2ul)
- Refer to BioBricks as 4D LacCass (for Lac inducible expression cassette), 1G ConsCass (for constitutive expression cassette), 11E VegRSac (for Constitutive promoter Pveg with RBS spoVG and SacB) for labelling.
- All the bands we need are on present on the 1G ConsCass and 4D LacCass, which means the digest is good for 3A Assembly. All we need now is the kerA and pSB1A3 ligation and the primers.
- We need to double-check the sizes of the insert and see if they are on the gel
Paper Work
Things to do:
- Figure out how to send things to sequencing
- Fill out expense sheet for ordering primers
- Order primers!! Kim and Arthur are out but the expense form needs to be filled out
- See Arthur and talk to him about paying the conference fees
- Finish coding website and upload to iGEM wiki source
Friday, August 16, 2013
- Need more AflII enzyme to do the pUB110 digestion.
- Tag: B.subtilisBackbone. pUB110 gel purification of samples 1 and 2 (from mini-prep) // double checking if enzymes are working ETHAN IS RUNNING THIS
- Tag: E.colikerAPlasmid. kerA to pSB1A3 (ampR plasmid) O.N. cultures of 3 picked colonies (mark with sharpie on back of petri with number to note which ones you’ve picked) not now, maybe Monday, read up on VF2/VR primers instead (Happens later)
- Tag: B.subtilisPromoter. Agar slab, re-streak
- Tag: E.colikerAPlasmid, B.subtilisBackbone. Using the columns that Ethan saved, he did gel extraction with warmed EB
- Tag: PaperWork.
- Figuring out how many of and of what samples to send to sequencing (3 samples of the kerA-pSB1A3 plasmid, 3 samples of linker-pUB110, if that ever gets done)
- Melissa leaves office by 4pm,
- Monday, we can send samples for sequencing (can we just hand them the plate--check the DNA core website for details).
- If someone can make a mailchimp campaign and start sending out requests for donations
- Isolated more pUB110: miniprep sample #3
Saturday, August 17, 2013
pUB110 linker and RBS agar stab, O.N. cultures of E. coli with kerA-pSB1A3, and Transformation of DH5a cells with RFP (for amplification)
- Alice put the linker (pre-fix and suffix ONLY for pUB110)+RBS (ribosome binding site) agar stab in the incubator.
- Annie made ampicillin stock 100mg/ml: Dissolved 5mg ampicillin in 50 µl of dH20. It’s in the -20ºC.
- Annie made 5 overnight cultures of kerA-pSB1A3 in 3ml LB and 3µl of amp stock (8/17 O.N. kerA-pSB1A3 #1-5). The colonies are labeled on the agar plate. Overnight cultures are put in the shaker in Nora’s Lab.
- Made and autoclaved 1L 1XLB. Ivan Poured 15 camR plates.
- Ivan and Alice transformed RFP into E coli. dh5α. (For purpose of amplification since we don’t have enough of it)
- Next Steps:
- Pick colonies from pUB110 linker-RBS agar stab and set up O.N. cultures
- Miniprep 3 of the O.N. kerA-pSB1A3 cultures
- Pick colonies from RFP transformation of DH5α E. coli
Week 4
Sunday, August 18, 2013
Restocking
- Poured 12 cam and amp plates each, using 250ml of each
- Note: when washing anything that will touch bacteria in the future, don’t use antimicrobial soap that’s provided. wash thoroughly with ddH2O instead.
Amount of antibiotics added:
| Amp 100ug/ml | added 0.025g/250ul Amp to 250ml LB-agar |
| Cam 25ug/ml | added 0.0063g/125ul Cam to 250ml LB-agar |
B.subtilis Backbone
- Ran an overnight digest of 8/16 pUB110 mp3 (8/18 pUB110 mp3 Digest)
- 7.5ul nuclease-free H2O
- 5ul pUB110 mp3
- 1.5ul 2.1 buffer
- 0.5ul AflII
- 0.5ul NdeI
- Incubated from 5:30pm - 10:30am
E.coli kerA Plasmid
- The overnight cultures set up yesterday showed no growth, perhaps because the caps were tightened too much.
- We set up three more overnight cultures (protocol) of 8/15’s kerA-pSB1A3 transformation (8/18 O.N. kerA-pSB1A3 #6-8)
- Each of the three colonies were put in 3 mL LB + 3 ul 50mg/ul ampicilin at 9:45 am
- the caps of 8/17’s O.N. kerA-pSB1A3 cultures #3, #4, #5 were loosened in hopes that the colonies will grow. 8/17’s O.N. kerA-pSB1A3 cultures #1, #2 were discarded because there were no pellets.
- The cultures were checked at 6pm. However, there was still no cell growth
- Perhaps the colonies were not Amp resistant, and we must make new ampicillin plates.
B.substilis Promoter
- Plate Pveg + RBS if there's growth.
- The pUB110 linker and RBS agar stab was incubated yesterday at 37C overnight but no growth is visible. The plate will continue to incubate until tomorrow. If there’s no growth by tomorrow morning, we must request another agar stab.
B.substilis RFP
- Made three overnight cultures of the RFP BioBrick in DH5a (8/18 O.N. RFP #1, #2, #3)
- Each colony was put in 3 ml LB + 1.5 ul of 50mg/ul chloramphenicol on a shaker at 37C at 11:30 am
Paper Work
- making binder full of necessary info and protocols
Next Steps:
- Make new ampicillin plates
- Re-do kerA-pSB1A3 transformation on new amp plates
Tuesday, August 20, 2013
kerA-pSB1A3 ligation and RFP digestion results
- Measured the concentration of the RFP digested yesterday
- [8/19 RFP Digest] = 74.7 ug/ul
- Ran a 2% gel of the RFP digest
- Lane 2: Ladder (8 ul)
- Lane 4: 8/19 RFP mp#2 Digest (2 ul) + loading dye (2 ul)
- Added 5/10 ul EtBr to the buffer
- There is only 1 band in lane 4.
- Email iGEM to request new Pveg+RBS construct purchased
- Ligated kerA and pSB1A3 with control experiments (Ivan)
- The ligations were set up as follows:
| uncut w/o ligase Control | cut w/ ligase Control | 3:1 | 5:1 | 10:1 | |
| Vol. Insert kerA digest 8/19 (uL) | N/A | N/A | 1 | 1.5 | 3 |
| Vol. Vector pSB1A3 digest (uL) | (18E)** 1 | 1 | 1 | 1 | 1 |
| Vol. Buffer (uL) | 1 | 1 | 1 | 1 | 1 |
| Vol. Ligase (uL) | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Vol. H2O (uL) | 8.5 | 7.5 | 6.5 | 6 | 5.5 |
used 18E (e. coli promoter from plate 5) instead of pSB1A3, has plasmid [http://parts.igem.org/File:PBca1020-r0040.jpg BBa_J61002] w/ AmpR
- The concentrations of the components of the ligations were as follows:
- kerA 8/19 digest: 90ng/ul
- pSB1A3 digest: 24ng/ul
- 18E miniprep: 963ng/ul
- The ligations incubated at RT overnight
- Another kerA-pSB1A3 ligation is being run in parallel in case the first does not work (Ethan):
- Incubated overnight at 4°C
- ligation reactions:
- ker A gel extraction (8/16) = 115 ng/ul (~1 kb)
- ker A gel extract (8/13) = 75 ng/ul
- pSB1A3 digest = 25 ng/ul (~ 2 kb)
| reaction | 1 | 2 |
| kerA | 1 ul kerA (8/16) = 115 ng | 2 ul kerA (8/13) = 150 ng |
| pSB1A3 | 1 ul pSB1A3 = 25 ng | 0.5 ul pSB1A3 = 12.5 ng |
| molar ratio (insert/vector) | 2.3 | 6 |
| 1 ul 10X ligation buffer | 1 ul 10X ligation buffer | |
| 0.5 ul ligase | 0.5 ul ligase | |
| 6.5 ul water | 6 ul water |
- Digested (Enzymatic Digestion Protocol) kerA overnight (Ethan)
- 10 ul kerA
- 2 ul digest buffer
- 0.5 ul EcoRI (F.D.)
0.5 ul SpeI (F.D.) 7 ul H2O
- Digested pSB1A3 overnight (Ethan)
- 5 ul pSB1A3 (linear)
- 1 ul digest buffer
- 0.5 ul EcoRI (F.D.)
- 0.5 ul SpeI (F.D.)
- 3 ul H2O
Wednesday, August 21, 2013
Transformation with kerA-pSB1A3 with control experiments and Transformation of B. subtilis WB700 with pUB110
- Transformed E. coli ([Transformation protocol]) with yesterday’s kerA-pSB1A3 ligation
- used 8/19 kerA digest (90ng/ul), the only pSB1A3 digest (24ng/ul), and 18E miniprep (963ng/ul)
- incubated overnight at 37C starting 2:30pm
- the following experiments, including controls, were performed
| plate/tube # | 1 | 2 | 3 | 4 (has small crack) | 5 |
| tube | 3:1 | 5:1 | 10:1 | cut vector w/ ligase - control | uncut vector w/o ligase - control |
| Insert (kerA digest 8/19) | 1ul | 1.5ul | 3ul | N/A | N/A |
| Vector (pSB1A3 digest) | 1ul | 1ul | 1ul | 1ul | (18E)** 1ul |
| Buffer | 1ul | 1ul | 1ul | 1ul | 1ul |
| Ligase | 0.5ul | 0.5ul | 0.5ul | 0.5ul | 0.5ul |
| H2O | 6.5ul | 6ul | 5.5ul | 7.5ul | 8.5ul |
used 18E (e. coli promoter from plate 5) instead of pSB1A3, has plasmid [http://parts.igem.org/File:PBca1020-r0040.jpg BBa_J61002] w/ AmpR
- About control experiments:
- Uncut vector (BBa_J61002) withOUT ligase: checks for viability of competent cells and antibiotic resistance of the plasmid, as well as transformation procedure itself
- expect colonies
- since we are using a linearized plasmid, is there another uncut vector we could use that has the same antibiotic resistance so we can check the competency of our cells?
- cut vector WITH ligase: shows background due to vector recircularization (and uncut vector)
- expect few colonies (few more than without ligase), since only the plasmids which did not get cut during the digest or the plasmids which did get cut but are able to recircularize with the ligase will be transformed
- this is the most important control. theoretically, there should be no colonies, but there is always some background, so as long as we have a lot more colonies on the vector with insert (actual ligation reaction) plate, it's okay.
- The following controls were not carried out but should be and are useful for future experiments:
- cut vector withOUT ligase: shows background due to uncut vector
- expect few colonies, since only the plasmids which did not get cut during the digest will be transformed
- only do this one if you have extra cut vector, if we don't set it up it's not a big deal, since cut vector with ligase will just show the total background due to uncut vector and vector recircularization.
- insert ONLY WITH ligase: checks for contamination of intact plasmid in ligation or transformation reagents
- expect no colonies, since if there are colonies it indicates contamination of intact plasmid carrying antibiotic resistance in reagents since only the insert was used which should not be able to recircularize into a construct which can be transformed
- this control is optional, but if we are concerned about contamination of reagents with plasmids this could help, so I think we should do it if there is extra insert
- More info on control ligation experiments can be found [http://www.addgene.org/plasmid_protocols/DNA_ligation/ here].
- Analysis of yesterday’s Agarose gel of digested RFP in lane 4:
- the fragment is about 2 kb long
- The BBa_E1010 insert (RFP) is 706bp long, while the vector is 2070 bp long.
- It seems like we have our vector present (the approximately 2kb band in lane 4) but not our insert.
- To check if the BioBrick actually has RFP, plate E. coli with the plasmid with IPTG.
- After checking the kit again, we realized that the BioBrick we isolated from plate 3 well 6G is BBa_R0080, not BBa_E1010.
- The insert in BBa_R0080 is 149 bp, which explains why we couldn’t see it on the gel because it was run off.
- Transformed B. subtilis (Transformation Protocol) with pUB110
- make O.N. cultures tomorrow in LB Medium
Thursday, August 22, 2013
Results of kerA-pSB1A3 transformation and control experiments and Transformation of E. coli with RFP
- Transformation Plate Results of kerA-pSB1A3
| plate/tube # | 1 | 2 | 3 | 4 (has small crack) | 5 |
| tube | 3:1 | 5:1 | 10:1 | cut vector w/ ligase - control | uncut vector w/o ligase - control |
| Insert (kerA digest 8/19) | 1ul | 1.5ul | 3ul | N/A | N/A |
| Vector (pSB1A3 digest) | 1ul | 1ul | 1ul | 1ul | (18E)** 1ul |
| Buffer | 1ul | 1ul | 1ul | 1ul | 1ul |
| Ligase | 0.5ul | 0.5ul | 0.5ul | 0.5ul | 0.5ul |
| H2O | 6.5ul | 6ul | 5.5ul | 7.5ul | 8.5ul |
| Colonies | >10 | >10 | >10 | 0 | 0* |
used 18E (e. coli promoter from plate 5) instead of pSB1A3, has plasmid [http://parts.igem.org/File:PBca1020-r0040.jpg BBa_J61002] w/ AmpR
- The positive control (Tube #5) did not work, even though there should have been colonies. Perhaps there were no colonies because the 18E tube was not actually being the miniprep. The concentration did seem too high.
- Set up three O.N. cultures of kerA-pSB1A3 E. coli in 3 ml LB with ampicillin
- Transformed E. coli with RFP BBa_E1010, well 12N, plate 3 (12N3)
- Followed PCR tube competent cell protocol
- Spread on plates with chloramphenicol and IPTG
Friday, August 23, 2013
Sending kerA-pSB1A3 Transformants for Sequencing and overnight cultures of RFP transformants
- Isolated Plasmid DNA by Miniprep of 8/18’s O.N. kerA-pSB1A3 cultures #4 and #6 and measured their concentrations to send for sequencing:
| colony | ng/ul |
| 4 | 150 ng/ul |
| 6 | 185 ng/ul |
- The university's DNA sequencing facility recommends that the concentration must be at least size (kb)/10 = [ug/ul]
- 3.35kb / 10 = 0.335ug/ul → 335ng/ul
- The concentrations of the kerA-pSB1A3 above are too low.
- Instead we sent the 3:1 ligation plate (see 8/22/13) for colony picking and sequencing of 5 samples.
- Primers VF2/VR, which are used for sequencing, need to be at 4uM
- dilution: (100uM)(2ul)=(4uM)(50ul) ⇒ 2ul primer in 48ul dH2O
- Results of yesterday’s RFP Transformation Plate:
- The colonies are not red. The transformation does not seem to have worked.
- After 5 hours of additional incubation 11am-4pm (20hrs total), white mold started growing on the plate.
- Plate is not cam resistant?
- Transformed E. coli again onto another cam plate, more recently made (agar plates protocol)
- RT SOC incubation 4:50 - 5:50
- Incubation on 37C shaker from 6 pm
- Set up two colonies from yesterday’s RFP transformation plate for overnight culture in 3 mL LB with chloramphenicol to double check if they are cam resistant colonies at 5:30pm.
Week 5
Monday, August 26, 2013
pUB110 digestions, keratinase assay, and quikchange mutagenesis research
B.subtilis Backbone
- Used FastDigest enzymes and green dye to digest pUB110 for 1 hour
- Ran the pUB110 digest (8/26 pUB110 digest with FastDigest enzymes) on a 1% agarose gel:
- lane 1: ladder (10ul)
- lane 2: fastdigest (20ul)
- The pUB110 digest does not seem to have worked.
- Will try again tomorrow or Wednesday
- Expected sizes:
- uncut pUB110: ~4500 bp
- vector: ~3500 bp
- AflII - NdeI: ~600 bp
- NdeI - NdeI: ~400 bp
B.substilis RFP
- Realized that BBa_E1010 has no promoter so we shouldn’t expect RFP expression anyway.
- Realized that we might not even need the RFP since the instructions for submitting a new biobrick backbone says we only need the prefix/suffix.
Keratinase Assay
- Finished compiling keratin plate/keratinase assay information
- Called Western Live Poultry and Alliance Poultry Farm to ask for chicken feathers. Will be stopping by tomorrow with garbage bags, gloves, and boots to pick up some really dirty feathers that we’ll need to wash before use.
Quikchange Mutagenesis
- For the optimization of keratinase activity
- Quikchange mutagenesis protocols to look at:
- http://openwetware.org/wiki/Site-directed_mutagenesis
- http://www.stanford.edu/~loening/protocols/Site_Directed_Mutagenesis.pdf (w/ Pfu)
- https://www.neb.com/products/e0554-q5-site-directed-mutagenesis-kit
- https://www.neb.com/protocols/2012/08/29/protocol-for-q5-high-fidelity-2x-master-mix-m0492
- Compiled a protocol for re-using cuvettes.
Tuesday, August 27, 2013
Troubleshooting pUB110 Digest
- FastDigest enzyme digest may not have worked because we used 1.5/2.1 buffer instead of FastDigest buffer
- We also used too much of the miniprep (5 ul instead of 1.5 ul)when we should have used less.
Wednesday, August 28, 2013
pUB110 digestion and B. subtilis electroporation
- Bacillus subtilis electroporation: following protocols stated below
- Grew wb700 overnight to OD600 = 4.1 and 7.4
- Started two 10 ml cultures (@ 12 pm) with
- OD600 = 4.1 (500 ul) and
- OD600= 1.3 (750 ul)
- x to OD600 = 0.2 (OD after dilution)
- 4.1 * (0.05) = .205 OD
- 1.3 * (0.075) = 0.0975 OD ???? too low
- subtilis doubling time = 25-40 minutes
- 1:30→ 0.4 abs
- 2:30→ 0.85 abs and 1.0 abs
- Electroporated at 5:30pm, following Eppendorf and its original “High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis” (Xue et al. 1999)
- Measured concentration of pBU110 miniprep: [pUB110 miniprep 1] = 367ng/ul sample
- Digested pUB110 overnight with FastDigest enzymes: incubated 6pm
- 1.5 ul pUB110
- 1.5 ul FastDigest buffer
- 0.5 ul AflII
- 0.5 ul NdeI
- 11 ul nuclease-free H2O
- Next Steps:
- Run 1% agarose gel of 8/28 pUB110 FastDigest and 1 ul of undigested miniprep
Thursday, August 29, 2013
pUB110 digest and Results of B. subtilis electroporation
- 8/28 pUB110 O.N. digest with FastDigest enzymes: incubated 6pm-11:30am
- 1.5ul (miniprep 1: 367ng/ul sample) pUB110
- 1.5ul FastDigest buffer
- 0.5ul AflII
- 0.5ul NdeI
- 11ul nuclease-free H2O
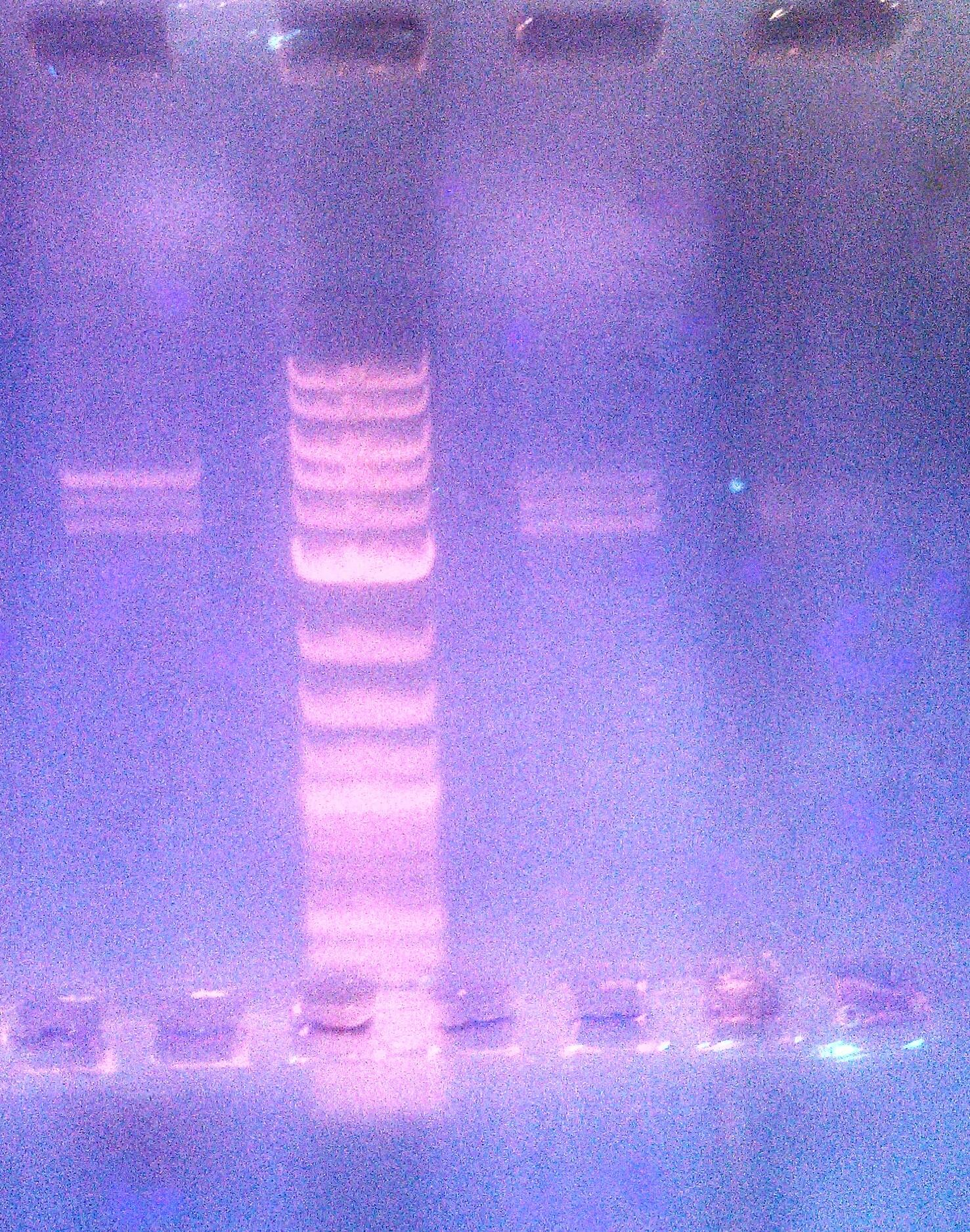
- Ran a 1% agarose gel of 8/28 pUB110 digest (11:45 am - at 120 V)
- lane 1: previously failed digest from 8/27 (5ul dye + 15ul sample)
- lane2: 2 log ladder (10ul)
- lane3: 8/28 pUB110 digest (15ul)
- lane4: miniprep pUB110 (1ul dye + 1ul sample)
- Gel purified (Qiagen gel extraction protocol) the lower two bands: grouped them together into tubes called Upper Band (UB) and Lower Band pUB110 (LB) digest
- Lane 1: Ladder
- Lane 2: 8/28 pUB110 digest
- Measured the concentrations with nanodrop:
- LB: 3.2ng/ul
- UB: 5.2ng/ul
- pUB110 re-digest (with FastDigest conditions, same as 8/26/13)
- used 2ul pUB110 sample with fastdigest conditions, incubated 1hr at 37C (total volume 20ul)
- measured concentrations of digest: 8.1 ng/ul
E.coli kerA Plasmid
- Troubleshooting kerA-pSB1A3 ligations and transformations:
- try using phosphatase to minimize self-ligations
- next digest: scale up 30ul volume, making sure to use water bath in heat block, do it with controls as well (single enzyme and etc)
Friday, August 30, 2013
kerA-pSB1A3 Sequencing Results
E.colikerAPlasmid
- We recieved our sequencing results of our kerA-pSB1A3 colonies
- When we analyzed the sequences for alignment with our kerA DNA sequence, it was apparent that our kerA biobrick was not ligated into the plasmid.
September
Week 1
Tuesday, September 3, 2013
Promoter+RFP transformation, kerA-pSB1A3 ligation and digestion, kerA digestion, pSB1A3 digestion
B.substilisRFP
- Transformed E. coli with BBa_J23100 (constitutive promoter family + RFP) for pSB1A3 backbone
- Followed transformation protocol for E. coli PCR tubes
- For making stock pSB1A3 backbone and for trying ligation without gel extraction
- Plates dried in the incubator, but [http://www.protocol-online.org/biology-forums-2/posts/6843.html bubbles at bottom] appeared, which could imply contamination
- Incubation at 37C overnight
- The following morning there were no colonies.
- Must try transformation again except with control experiments.
E.coli kerA Plasmid
- Perhaps we should treat the plasmid with antarctic phosphatase to prevent self-ligation
- Digested kerA and pSB1A3 (Ezymatic Digestion Protocol) overnight at 37°C
| Reaction | 1 | 2 |
| 6 ul linearized pSB1A3 backbone = 150 ng | 2.5 ul kerA (408 ng/ul) = 800 ng DNA | |
| 1.5 ul buffer | 1.5 ul buffer | |
| 0.5 ul EcoRI | 0.5 ul EcoRI | |
| 0.5 SpeI | 0.5 SpeI | |
| 7.5 ul water | 7.5 ul water |
- Next Steps:
- kerA-pSB1A3 ligations and transformation of E. coli
- Start O.N. cultures of RFP transformations and cut amp concentration to 50 ug/ul
Wednesday, September 4, 2013
Results of RFP Transformation, kerA-pSB1A3 ligations and transformation of E. coli, and testing transformation efficiency of E. coli cells
E.coli kerA Plasmid
- Ligated yesterday’s digested kerA and pSB1A3 overnight at 4°C (Ligation Protocol)
- 9 ul kerA digest (40 ng/ul)
- 1 ul pSB1A3 digest (12.5 ng/ul)
- 1.5 ul buffer
- 0.5 ul ligase
- 3 ul water
B.substilis RFP
- BBa_J23100 RFP transformation resulted in no colonies. Must try again.
- Other biobricks with AmpR plasmids we can try transformations with:
- J23119 18A, plate 5 (in pSB1A3)
- J23100 18C, plate 5 re-transform (in BBa_J61002)
- [http://parts.igem.org/Part:BBa_J23101 J23101] 18E, plate 5 (in BBa_J61002)
kerA BioBrick Troubleshooting
- Perhaps, there is something wrong with our kerA?
- Redid kerA biobrick PCR (gibson assembly with geneblocks)
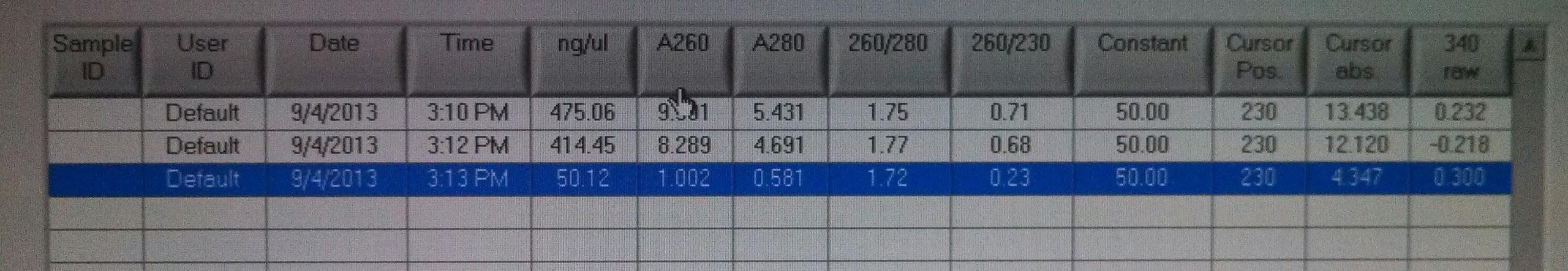
- Measured concentrations of kerA biobrick PCR with nanodrop:
- kerA biobrick PCR #1: 475ng/ul
- kerA biobrick PCR #2: 414ng/ul
- kerA digest 8/21: 50ng/ul
- Tested transformation efficiency of our competent cells with iGEM’s [http://parts.igem.org/Help:Transformation_Efficiency_Kit Transformation Efficiency Kit]: using 5pg/ul, 10pg/ul, 20pg/ul
- Dephosphorylated restriction digests using antarctic phosphorylase:
- 15 ul kerA/pSB1A3 digest (9/3)
- 1.7 ul buffer
- 0.5 ul phosphorylase
- Incubated at 37°C (3:30-4:00 pm)
- Heat inactivated enzyme at 70°C (5 minutes)
- Ligated dephosphorylated pSB1A3 and kerA digests and incubated overnight at 4C
| 1 | 2 | 3 |
| 2 ul pSB1A3 Digest (18 ng) | 2 ul pSB1A3 Digest (18 ng) | 2 ul pSB1A3 Digest (18 ng) |
| 4 ul kerA (188 ng) | 8 ul kerA | 1 ul buffer |
| 1 ul buffer | 1.05 ul buffer | 0.5 ul digest |
| 0.5 ul ligase | 0.5 ul ligase | 6.5 ul H2O |
| 2.5 ul H2O |
- Set overnight culture (protocol) of B. subtilis for electroporation tomorrow
- Autoclaved materials and tips and Made more chloramphenicol plates (protocol)
- Transformed kerA-pSB1A3 ligation into DH5Alpha E. coli
- Following PCR tube transformation protocol
- 1ul DNA into thawed competent cells on ice per tube (3 sets of 3 tubes)
- Incubate set #1, 2 on ice for 30min, set #3 on ice for 10min
- Heat shock in PCR machine 45s at 42C
- Recover on ice for 5min
- Add to 1mL LB and incubate on shaker for 37C for 1hr
- Spin down at 8rpm for 30s and plate on Cam plates
Thursday, September 5, 2013
Results of Transformation efficiency, B. subtilis electroporation (2nd Trial), and RFP transformation
- Results of RFP Control with Transformation Efficiency Kit testing PCR competent cells
- Negative Control Plage
- There weren't any colonies on the negative plate, and we got colonies on the positive plates.
- We can conclude that the competent cells worked.
- However, the number of colonies from plate to plate for the same concentrations weren't consistent. That may be due to inconsistent volume of competent cells in the PCR tubes.
- Electroporation of B. subtilis WB700 (2nd Trial): following "High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis" (Xue et al. 1999)
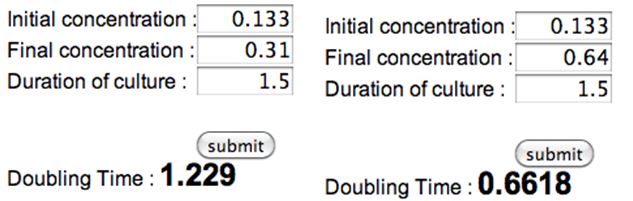
- Calculating Doubling Time Constant
- Average doubling time = 1.156
- ~69min doubling time from 8/28 OD measurements, using the 4.1OD → 0.205OD → 0.85OD or 1OD, taking ave
- Doubling time (formula from wikipedia)
N(t) = 0.85abs C = 0.133abs (see below) d = 1.156 t = 1.156 * [log(.85/.133)/log2] = 3.09 hrs
- rough estimate of doubling time to double check above figure:
- need for 0.9nm OD
- 3.46nm/26ml
- starting OD calculated at 0.133 (w/o dilution)
- x2 = 0.266
- x2 = 0.532
- x2 = 1.064 ~roughly
O/N culture OD (dilution): 0.346nm (1:10 dilution in water, 100ul in 900ul water) original OD in 15ml tube is 3.46nm
Diluted Initial OD (w/o dilution): 0.133nm (1ml each in 25ml sorbitol and 25ml LB)
Desired OD (w/o dilution): 0.85nm
| 0.5M Sorbitol in LB, 25 mL | LB, 25 mL | |
| 10am-11:30am | 0.133 nm | 0.133 nm |
| 11:30 am | 0.031*10 = 0.31 nm | 0.064*10 = 0.64 nm |
| 12-12:20 pm | N/A | 0.104nm*10 = 1.04 nm (used Ethan's spec) |
| 12-1:35pm | 0.111nm*10 = 1.11 nm (used Nora's spec) | N/A |
LB only
N(t) = 0.85abs
C = 0.64abs (see below)
d = 0.662
t = 0.662 * [log(.85/.64)/log2] = 0.27 *60min = 16min → 20min
[copy, paste into google search] log(.85/.64)/log(2) *0.662
LB 0.5M sorbitol
N(t) = 0.85abs
C = 0.31abs (see below)
d = 1.229
t = 1.229 * [log(.85/.31)/log2] = 1.79 * 60min = 1hr 47min → 1hr 50min
[copy, paste into google search] log(.85/.31)/log(2) *1.229
B. subtilis Electroporation Protocol from Xue et al. 1999:
- Cool cells in 50ml Falcon tubes in ice-water, 10min
- Spin down 3,000rpm for 5min (Nora’s lab)
- Resuspend in 650ul electroporation medium and transfer to 1.5ml tubes
- Spin down 5,000xg for 5min (BSLC lab)
- Wash 3x with electroporation medium (500ul)
- Resuspend in 1/40 of culture volume of electroporation medium
- resuspend in 26/40 = 650ul
- dilution pUB110 #1 244ng/ul: (244ng/ul)(x ul) = (10ng/ul)(200ul) → x = 8.2ul
in 0.1cm gap
Re-using Cuvettes: (protocol) 500ul H2O mix up and down 3x, EtOH mix up and down with 10ul pipette, make sure you see flakes of dead cells on the first mixing, 2x, pour EtOH out as much as possible and air dry for 10min, wipe cap with EtOH too.
| number | contains | kV | time constant | notes |
| 1 | LBS + DNA | 2.0 | 4.08 | no arc |
| 2 | LB + DNA | 2.0 | N/A | didn't wash after initial use, arc'd, immediately filled with EtOH and set aside |
| 3 | LBS + DNA | 2.1 | 3.96 | new cuvette |
| 4 | LBS + DNA | 2.0 | 1.70 | incubated sample to see what hapens; washed with 1mL H2O 3x, lots of EtOH 2x, dried out carefully, on plain plate |
| 5 | LBS + DNA | 2.0 | 4.16 | no arc |
| 6 | LB + DNA | 2.1 | 3.06 | washed 2x H2O instead of 3x, wasn't as careful, arc'd, incubated |
| 7 | LB + DNA | 2.1 | 4.2 | washed carefully, no arc |
| 8 | LB (N) | 2.1 | 4.2 | washed carefully, no arc |
Incubated tubes at 6:40pm-9:40pm. Incubated plates at 10pm.
- Plated on 50ug/ml Kan plates → may be issue since protocol recommends 20ug/ml Kan plates?
Friday, September 6, 2013
Results of B. subtilis electroporation and kerA-pSB1A3 ligations (all trials)
E.coli kerA Plasmid
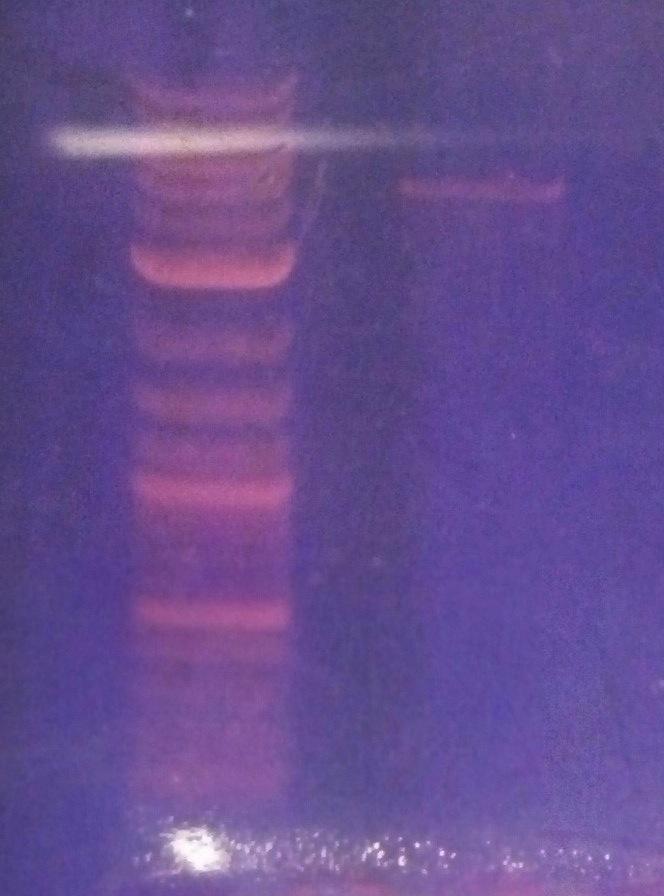
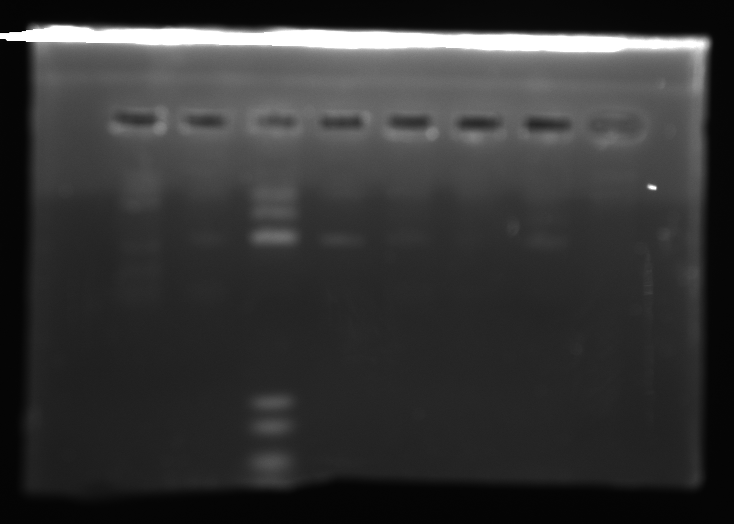
- Ran a 1% agarose gel of kerA ligations to see if the ligations actually worked
- lane1: ladder (10ul)
- lane2: kerA pcr (0.5ul of 457ng/ul in 5ul)
- lane3: kerA-pSB1A3 ligation 8/15(5ul) [ looks like there some ligation product, but transformation didn’t work ]
- lane4: cut vector w/ ligase 8/20 (8ul)
- lane5: 3:1 ligation 8/20 (8ul)
- lane6: 5:1 ligation 8/20 (8ul)
- lane7: 10:1 ligation 8/20 (8ul)
- kerA-pSB1A3 ligation digest 8/20 (3:1 ratio tube)
- kerA size: ~1199 bp
- pSB1A3 size: ~2155 bp
- expected size: = ~3354 bp
B.subtilis Backbone
- Made overnight cultures of B. subtilis BD 366 in 3 mL LB with kanamycin to isolate pUB110 (from 7pm - 1pm)
- B. subtilis Electroporation plates:
- There were no colonies on the negative plate, while there is only one colony on the other plates. It was incubated for longer, and restreaked the following day.
- Transformed (protocol) E. coli with (three) kerA-pSB1A3 ligations and (two) RFP biobrick
- incubated 30 min on ice
- plated on Amp plates
Week 2
Monday, September 9, 2013
O.N. cultures of past RFP biobricks, kerA-pSB1A3 ligations, and electroporations; Prolonged incubation of electroporations; and Re-streaking RFP biobricks, electroporations, and most recent kerA-pSB1A3 ligation
- At 5pm set up O.N. cultures of:
| cam: RFP 8/22 | overnight culture of a RFP biobrick in pSB1C3 backbone |
| amp: RFP1 9/6 | overnight culture of a RFP biobrick in non-pSB1A3 backbone |
| amp: RFP2 9/6 | overnight culture of a RFP biobrick in pSB1A3 backbone |
| amp: 10:1 ligation #1 | old ligation plates, from 8/20, colony 1 |
| amp: 10:1 ligation #2 | old ligation plates, from 8/20, colony 2 |
| amp: 5:1 ligation #1 | old ligation plates, from 8/20 |
| amp: 5:1 ligation #2 | old ligation plates, from 8/20 |
| amp: 9/6 L2 | Ethan’s most recent ligation pSB1A3-kerA, only plate with colonies |
| kan: AY #3 EP #1 | one of my electroporation plates, colony 1 |
| kan: AY #3 EP #2 | one of Alice's electroporation plates, colony 2 |
| kan: EB N EP | Ethan’s electroporation plates, negative control |
| kan: EB 10ng EP #1 | Ethan’s electroporation plates, negative control, used 10ng of plasmid DNA, colony 1 |
| kan: EB 10ng EP #2 | Ethan’s electroporation plates, negative control, used 10ng of plasmid DNA, colony 2 |
| kan: EB 10ng EP #3 | Ethan’s electroporation plates, negative control, used 10ng of plasmid DNA, colony 3 |
| kan: EB 50ng EP #1 | Ethan’s electroporation plates, negative control, used 50ng of plasmid DNA, colony 1 |
| kan: EB 50ng EP #2 | Ethan’s electroporation plates, negative control, used 50ng of plasmid DNA, colony 2 |
| kan: EB 50ng EP #3 | Ethan’s electroporation plates, negative control, used 50ng of plasmid DNA, colony 3 |
| kan: EB 4x EP | Ethan’s electroporation plates, most recent plate that had colonies - did four overnight cultures |
- Incubated the following plates longer from 10:30 am and checked again at 4 pm:
| kan: AY #1 EP | one of Alice’s electroporation plates, wanted to see if more colonies would grow |
| kan: AY #3 EP | one of Alice’s electroporation plates, wanted to see if more colonies would grow |
| kan: EB 10ng EP | Ethan’s electroporation plates, wanted to see if more colonies would grow |
| kan: EB 50ng EP | Ethan’s electroporation plates, wanted to see if more colonies would grow |
| kan: EB N EP | Ethan’s electroporation plates, wanted to see if more colonies would grow |
- Restreaked and incubated until 9pm:
| amp: RFP1 9/6 | From BBa_J23100, non-pSB1A3 backbone, from Chloe’s RIBS class’ BioBrick used, original plate was messy plating |
| amp: RFP2 9/6 | From BBa_J23119, pSB1A3 backbone, from BioBrick kit, original plate was messy plating |
| kan: AY #3 EP | plate number 3, my electroporation plate with colonies; replated to try to get more colonies |
| amp: 9/6 L2 | Ethan’s most recent ligation pSB1A3-kerA, only plate with colonies |
- The feathers we ordered are here! Must pick them up tonight and we can make our feather keratinase assay.
- Made chloramphenicol plates 250 mL in total (protocol)
- Ligated kerA and antarctic phosphatase treated pSB1A3 and incubated overnight at 4°C (protocol):
| 1 | 2 | 3 | 4 | 5 |
| 1 ul pSB1A3 Digest (9 ng) | 1 ul pSB1A3 digest (9 ng) | 1 ul pSB1A3 digest (9 ng) | 1 ul pSB1A3 digest (9 ng) | 1 ul pSB1A3 digest (9 ng) |
| 0.5 ul ul KerA (220 ng) | 8 ul KerA (188 ng) | 1.05 ul buffer | 1 ul buffer | 1 ul buffer |
| 1 ul buffer | 1 ul buffer | 0.5 ul digest | 0.5 ul digest | 0.5 ul digest |
| 0.5 ul ligase | 0.5 ul ligase | 6.5 ul h2o | 6.5 ul h2o | 6.5 ul h2o |
| 2.5 ul h2o
total DNA = 22 ng/ul |
Tuesday, September 10, 2013
Miniprep of 9/09’s O.N. cultures
- Isolated plasmid DNA of yesterday’s O.N. cultures (see below) by miniprep
| cam: RFP 8/22 | overnight culture of a RFP biobrick in pSB1C3 backbone |
| amp: RFP1 9/6 | overnight culture of a RFP biobrick in non-pSB1A3 backbone |
| amp: RFP2 9/6 | overnight culture of a RFP biobrick in pSB1A3 backbone |
| amp: 10:1 ligation #1 | old ligation plates, from 8/20, colony 1 |
| amp: 10:1 ligation #2 | old ligation plates, from 8/20, colony 2 |
| amp: 5:1 ligation #1 | old ligation plates, from 8/20 |
| amp: 5:1 ligation #2 | old ligation plates, from 8/20 |
| amp: 9/6 L2 | Ethan’s most recent ligation pSB1A3-kerA, only plate with colonies |
| kan: AY #3 EP #1 | one of Alice's electroporation plates, colony 1 |
| kan: AY #3 EP #2 | one of my electroporation plates, colony 2 |
| kan: EB N EP | Ethan’s electroporation plates, negative control |
| kan: EB 10ng EP #1 | Ethan’s electroporation plates, negative control, used 10ng of plasmid DNA, colony 1 |
| kan: EB 10ng EP #2 | Ethan’s electroporation plates, negative control, used 10ng of plasmid DNA, colony 2 |
| kan: EB 10ng EP #3 | Ethan’s electroporation plates, negative control, used 10ng of plasmid DNA, colony 3 |
| kan: EB 50ng EP #1 | Ethan’s electroporation plates, negative control, used 50ng of plasmid DNA, colony 1 |
| kan: EB 50ng EP #2 | Ethan’s electroporation plates, negative control, used 50ng of plasmid DNA, colony 2 |
| kan: EB 50ng EP #3 | Ethan’s electroporation plates, negative control, used 50ng of plasmid DNA, colony 3 |
Wednesday, September 11, 2013
pUB110 digestion and gel analysis
- digested pUB110 minipreps with Nde I to test if electroporation was successful
- Ran a 1% agarose gel of the digest
- The DNA was a smear!!!! (genomic DNA?)
- The pUB110 minipreps were unsuccessful.
Thursday, September 12, 2013
Gibson Assembly of kerA, pUB110 ligation with linker (suffix and prefix), and Transformation of E. coli with 9/09 kerA-pSB1A3 ligations
- Ran a PCR 1:10 gibson assembly of kerA
- 1 ul prefix primer
- 1 ul suffix primer
- 2.5 ul buffer
- 1 ul mgcl2
- 0.5 ul polymerase
- 1 ul dntps
- 17 ul h2o
- → annealing temperatures (55°C, 58°C, & 62°C)
- Ligated pUB110 digest with gel purified linker (pUB110 suffix and prefix): Ligation protocol
- Concentrations of pUB110 upper band, pUB110 lower band, and linker
- linker gel extract = 115 ng/ul = 2.64 pmol/ul = 2640 femtomoles
- 0.5 microliters of linker = 57 ng
- The pUB110 linker is 67 b.p.
- pUB110 extract LB: 3.2ng/ul (8/29) = 1.1 fmol/ul
- pUB110 extract UB: 5.2ng/ul (8/29) = 1.7 fmol/ul
- pUB110 is 4500 kb
- Final amounts used:
- 10 ul of LB and UB = (11 fmol and 17 fmol, respectively)
- linker should be ~ 50 fmol (2 ul of 1:100 dilution)
- Transformed E. coli with 9/09 kerA-pSB1A3 ligations #1 - #5 (Transformation protocol)
Friday, September 13, 2013
Results of kerA gibson assembly and kerA ligation into pGEM-T Easy Vector
- The PCR of kerA was successful.
- Ligated kerA into [http://www.promega.com/resources/protocols/technical-manuals/0/pgem-t-and-pgem-t-easy-vector-systems-protocol/?__utma=1.1998193980.1380160105.1380160105.1380160105.1&__utmb=1.1.10.1380160105&__utmc=1&__utmx=-&__utmz=1.1380160105.1.1.utmcsr=google|utmccn=%28organic%29|utmcmd=organic|utmctr=%28not%20provided%29&__utmv=-&__utmk=166957246 pGEM-T Easy Vector] for eventual transformation into E. coli
(ng of vector × kb size of insert)/(kb size of vector) × insert:vector molar ratio = ng insert
(25 ng vector x 1 kb insert)/(3 kb vector) x 1:1 = 8.3 ng of insert = ~1.5 ul of PCR
| 1 | 2 | 3 | 4 | 5 | 6 |
| 1 ul buffer | 1 ul buffer | 1 ul buffer | 1 ul buffer | 1 ul buffer | 1 ul buffer |
| 0.5 ul pgem | 0.5 ul pgem | 0.5 ul pgem | 0.5 ul pgem | 0.5 ul pgem | 0.5 ul pgem |
| 1.5 ul PCR 1 (9/12) | 1.5 ul PCR 2 (9/12) | 1.5 ul PCR 3 (9/12) | 0.5 ul PCR 3 (9/12) | 3.0 ul PCR 3 (9/12) | 5.0 ul PCR 3 (9/12) |
| 0.5 ul ligase | 0.5 ul ligase | 0.5 ul ligase | 0.5 ul ligase | 0.5 ul ligase | 0.5 ul ligase |
| 6.5 ul h2o | 6.5 ul h2o | 6.5 ul h2o | 7.5 ul h2o | 5.0 ul h2o | 3.0 ul h2o |
Saturday, September 14, 2013
B. subtilis electroporation with control experiments
- Note: on re-using Cuvettes:
- 500ul H2O mix up and down 3x,
- EtOH mix up and down with 10ul pipette, make sure you see flakes of dead cells on the first mixing, 2x,
- pour EtOH out as much as possible and air dry for 10min, wipe cap with EtOH too.
- Electroporation Experiments:
| number | contains | kV | time constant | notes |
| 1 | neg control | 2.1 | 3.88 | no arc |
| 2 | LB + 50ng DNA | 2.1 | 4.20 | no arc |
| 3 | UB + 50ng DNA | 2.1 | 4.00 | no arc |
| 4 | positive control | 2.1 | 3.54 | no arc |
| 5 | LB + 100ng DNA | 2.1 | 3.96 | no arc |
| 6 | UB + 100ng DNA | 2.1 | 3.70 | no arc |
- Negative control: 6ul of dH2O
- LB Lig: 760ng/ul, diluted 1ul DNA w/ 14.2ul H2O, then added 1ul for 50ng, and 2ul for 100ng
- UB Lig: 690ng/ul, diluted 1ul DNA w/ 12.8ul H2O, then added 1ul for 50ng, and 2ul for 100ng
- Positive control: added 5ul, pUB110, 10ng/ul
Week 3
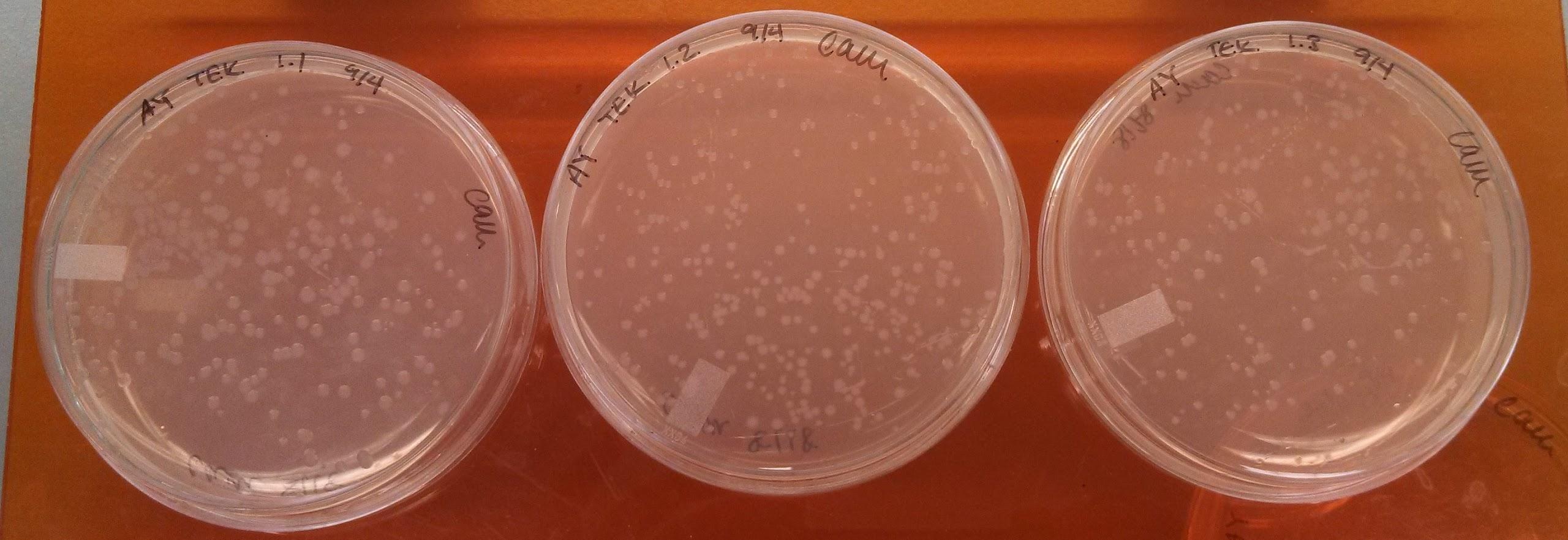
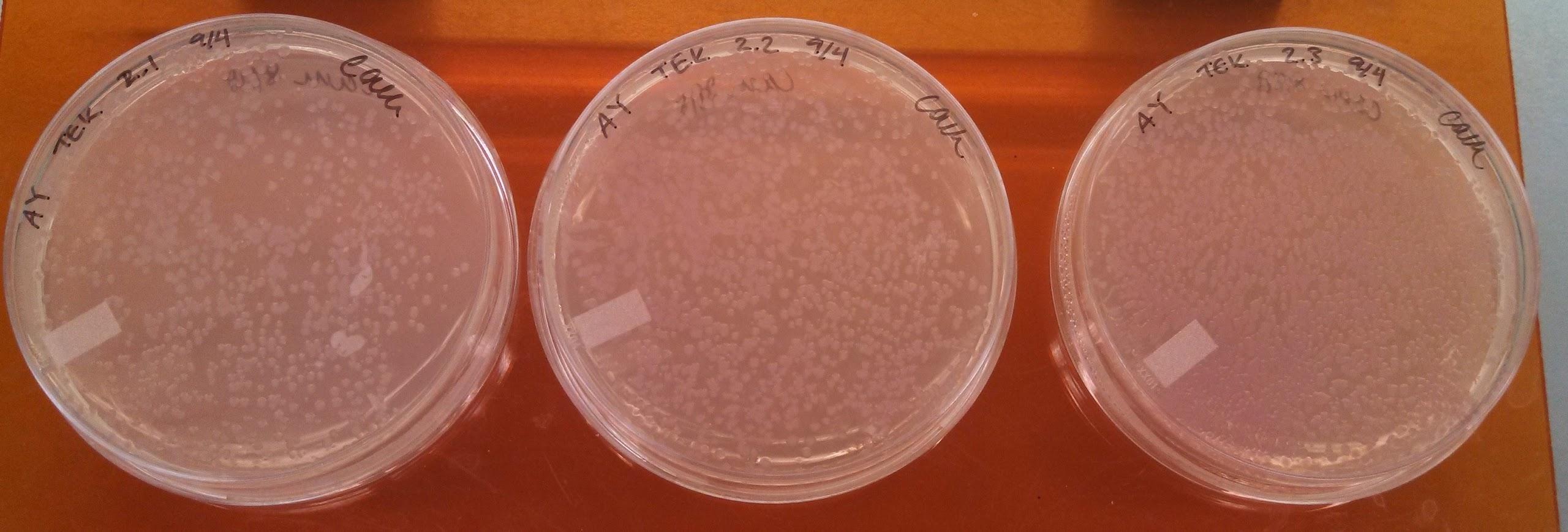
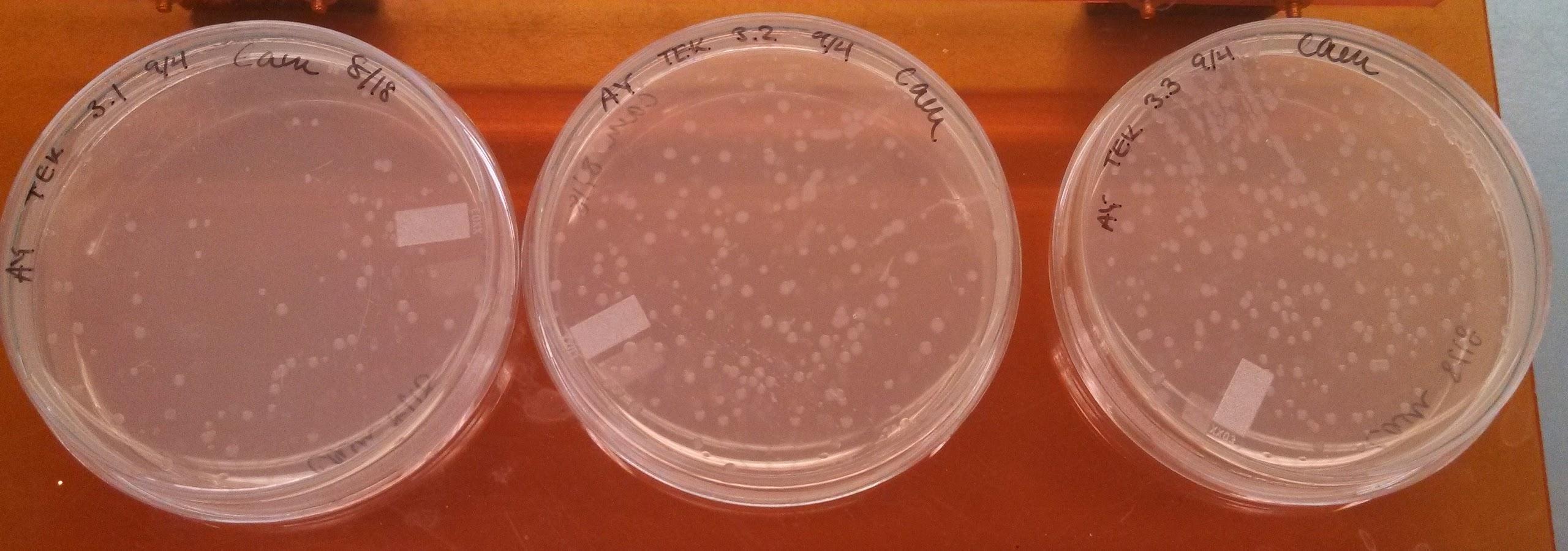
Monday, September 16, 2013
Results of Electroporation experiments with controls
- Results of yesterday’s electroporation experiments:
| Positive control | 2 colonies + a smear of E coli |
| Negative control | 1 colony |
| UB 50 | no distinct colonies, but some bacterial growth |
| UB 100 | no distinct colonies, but some bacterial growth |
| LB 50 | ~10 colonies |
| LB 100 | 1 colony |
- The electroporations seem to have not worked.
- Also, this may be insignificant, but there is a cam plate labeled “cam 9/9 AY + spmol tube 6” with 2-3 red colonies on it.
- The positive plates were put in the fridge.
 "
"