04/09/13
From 2013.igem.org
(Difference between revisions)
| Line 14: | Line 14: | ||
==Transformation results== | ==Transformation results== | ||
| - | From the above results,colonies were found growing on the resistance control plate, which indicates that our competent cells already express ampicillin resistance, but this is not meant to be so. Hence in order to confirm that our results are reliable | + | From the above results,colonies were found growing on the resistance control plate, which indicates that our competent cells already express ampicillin resistance, but this is not meant to be so. |
| + | *Hence in order to confirm that our results are reliable; | ||
| + | *colonies would be randomly selected from each of the two plates (20ul and 200ul) for the 3 TOD genes and | ||
| + | * cultured in luria broth + Amp. This broth would be incubated overnight in a 37 degrees shaker incubator. | ||
==Gel Electrophoresis== | ==Gel Electrophoresis== | ||
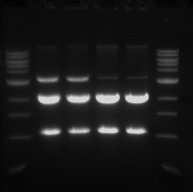
*The double digest (pSB1C3) from yesterday was run on a 1% gel. | *The double digest (pSB1C3) from yesterday was run on a 1% gel. | ||
Revision as of 12:34, 6 September 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Transformation results
From the above results,colonies were found growing on the resistance control plate, which indicates that our competent cells already express ampicillin resistance, but this is not meant to be so.
- Hence in order to confirm that our results are reliable;
- colonies would be randomly selected from each of the two plates (20ul and 200ul) for the 3 TOD genes and
- cultured in luria broth + Amp. This broth would be incubated overnight in a 37 degrees shaker incubator.
Gel Electrophoresis
- The double digest (pSB1C3) from yesterday was run on a 1% gel.
- This was done in order to confirm that the digest worked and also to purify the pSB1C3 vector.
- From left to right
- Sample 1.1 - track 2
- Sample 1.2 - track 3
- Sample 2.1 - track 4
- Sample 2.2 - track 5
- The 2 Kb bands are the pSB1C3 backbone
- The 1 Kb bands are the RFP biobrick
The two higher bands in tracks 2 and 3 are plasmids that were not digested.
Gel purification of the pSB1C3 backbone and RFP biobrick
- The Zymoclean Gel DNA recovery Kit was used to perform the purification and its protocol was followed.
- However the elution step was changed to 12ul of elution buffer, and this step was repeated.
Nanodrop of the samples
| Sample | Volume | Concentration ng/ul | 260/280 | 260/230 |
| pSB1C3 1 | 19.8 | 31.9 | 1.83 | 1.75 |
| pSB1C3 2 | 20 | 34 | 1.67 | 0.92 |
| RFP 1 | 20 | 28.8 | 1.57 | 0.71 |
| RFP 2 | 18.8 | 22.2 | 1.79 | 1.84 |
Double digest of TOD genes PCR products
Samples A (TodX), C (TodF) and E (ToBG) were digested with the enzymes XbaI and SpeI.
- Protocol
| Samples | A | C | E |
| Volume (ul) | 20 | 23.5 | 21 |
| SpeI (ul) | 1 | 1 | 1 |
| XbaI | 0.5 | 0.5 | 0.5 |
| Cutsmart Buffer | 6 | 6 | 6 |
| 5mTris Hcl (ul) | 32.5 | 29 | 31.5 |
- The samples were incubated at 37C for 90 minutes
- Heat kill the enzymes by incubating the samples at 80C for 20 minutes
Ligation of the TOD genes (X, F and ToBG) to the pSB1C3 backbone
The pGEM-T Vector System was used, However changes were made to the protocol. The promega biomath caculator (www.promega.com/biomath) was used to work out the ratio of insert to vector. The DNA concentration of the TOD genes can be found at Lab work 28/09/2013. The DNA concentration of the pSB1C3 and the RFP biobrick can be found above. The amount of vector DNA was added at 50ng.
Protocol
| Samples | A (TodX) | C (TodF) | E (ToBG) | RFP-sample 2 (positive control) | Background control |
| 2X Rapid ligation buffer (ul) | 5 | 5 | 5 | 5 | 5 |
| vector (pSB1C3-sample1) (ul) | 1.6 | 1.6 | 1.6 | 1.6 | 1.6 |
| T4 DNA ligase (ul) | 1 | 1 | 1 | 1 | 1 |
| PCR product (ul) | 2.3 | 1.4 | 3.1 | 3.6 | - |
| Deionized water for the final volume of 12 ul | 2.1 | 3 | 1.3 | 0.8 | 4.4 |
The reaction were incubated overnight at 4C for the maximum number of transformants
 "
"