10/09/13
From 2013.igem.org
(→Results of the Gel electrophoresis) |
(→Dissolution of polystyrene) |
||
| Line 14: | Line 14: | ||
==Dissolution of polystyrene== | ==Dissolution of polystyrene== | ||
| - | * | + | *a 20ml solution of 50% Limonene was made to determine if the dissolution of polystyrene from [[09/09/13]] would occur faster. |
| + | *as predicted, the 8cm x 5cm x 2cm PS block was completely dissolved in 9 minutes. | ||
==Gel electrophoresis== | ==Gel electrophoresis== | ||
Revision as of 15:15, 12 September 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Dissolution of polystyrene
- a 20ml solution of 50% Limonene was made to determine if the dissolution of polystyrene from 09/09/13 would occur faster.
- as predicted, the 8cm x 5cm x 2cm PS block was completely dissolved in 9 minutes.
Gel electrophoresis
- The samples from the double digest on 09/09/13 were run to confirm if the digestion worked.
- From the results, we would also be able to distinguish between cells that have been successfully transformed with the ligated pSB1C3/TOD genes from those with the recircularised pSB1C3.
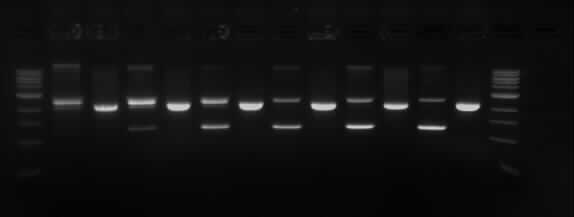
Results of the Gel electrophoresis
TODX Gel
Wells order: TODX1 (SpeI/XbaI), TODX1 (EcoRI/PstI); TODX2 (SpeI/XbaI), TODX2 (EcoRI/PstI); TODX3 (SpeI/XbaI), TODX3 (EcoRI/PstI); TODX4 (SpeI/XbaI), TODX4 (EcoRI/PstI); TODX5 (SpeI/XbaI), TODX5 (EcoRI/PstI); TODX6 (SpeI/XbaI), TODX6 (EcoRI/PstI).
The expected results were that the we would see the TODX genes drop off in separate band in all the digestions in its respective size (600bp). However as it can be seen in the picture above only the samples cut with SpeI and XbaI had two fragments and they are the wrong size (~ 1Kb). The samples TODX (1, 2 and 6) were sent to be sequenced before the digestions and it showed that the pSB1C3 plasmid re-circularized. This is because the two enzymes have complementary sticky ends.
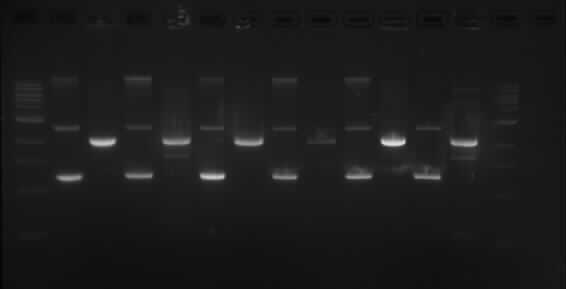
TODF
Wells order: TODF1 (SpeI/XbaI), TODF1 (EcoRI/PstI); TODF2 (SpeI/XbaI), TODF2 (EcoRI/PstI); TODF3 (SpeI/XbaI), TODF3 (EcoRI/PstI); TODF4 (SpeI/XbaI), TODF4 (EcoRI/PstI); TODF5 (SpeI/XbaI), TODF5 (EcoRI/PstI); TODF6 (SpeI/XbaI), TODF6 (EcoRI/PstI).
This shows the same results as TODX above.
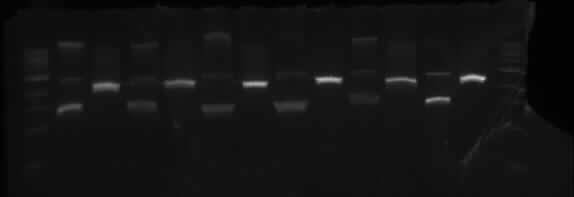
TOBG
Wells order: TOBG1 (SpeI/XbaI), TOBG1 (EcoRI/PstI); TOBG2 (SpeI/XbaI), TOBG2 (EcoRI/PstI); TOBG3 (SpeI/XbaI), TOBG3 (EcoRI/PstI); TOBG4 (SpeI/XbaI), TOBG4 (EcoRI/PstI); TOBG5 (SpeI/XbaI), TOBG5 (EcoRI/PstI); TOBG6 (SpeI/XbaI), TOBG6 (EcoRI/PstI).
This shows the same results as TODX above.
 "
"