From 2013.igem.org
Isolate plasmid from overnight culture
- Using Omega mini prep kit protocol
- Samples:
- TODF 1,2,3,
- TODX 1, 2, 3
- TOBG 1, 2, 3,
- RFP 1, 2, 3
| sample | Volume (ul) | Concentration (ng/ul)
|
| TODF1 | 43 | 120
|
| TODF2 | 43 | 98.8
|
| TODX1 | 42 | 399.3
|
| TODX2 | 44 | 442.2
|
| TODX3 | 43 | 417.5
|
| TOBG1 | 43 | 262.4
|
| TOBG2 | 47 | 319.2
|
| TOBG3 | 41 | 338.4
|
| RFP1 | 41 | 77.7
|
| RFP2 | 41 | 32.7
|
| RFP3 | 42 | 59
|
Glycerol stocks
- Took 750ul from overnight culture and centrifuged at full speed for ~2 minutes
- Supernatant was removed
- Pellet resuspended in 375ul of HMFM
- Samples were then frozen at -80
Gel Purification of the pGEM-T pasmid double digestions ran in the 11/09/13
The ge was purified by using the The Zymoclean Gel DNA recovery Kit and its protocol was followed.
Nanodrop Results
| Sample | Volume | Concentration (ng/ul) | 280/260 | 260/230
|
| TODX | 48.5 | 7.1 | 1.83 | 0.72
|
| TODF | 45.5 | 21.7 | 1.89 | 1.19
|
| TOBG | 47.7 | 9 | 1.77 | 0.70
|
| pSB1C3 (RFP) | 46.2 | 7.3 | 1.81 | 0.16
|
Digestion of the samples purified with the enzyme PstI-HF
| DNA volume | Volume | Cutsmart buffer | Enzyme volume | dH2O added to total volume of 60 ul
|
| TODX | 23 | 6 | 1 | 30
|
| TODF | 22.5 | 6 | 1 | 30.5
|
| TOBG | 22 | 6 | 1 | 31
|
| pSB1C3 (RFP) | 20.5 | 6 | 1 | 32.5
|
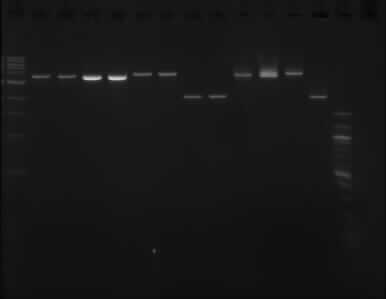
The picture above shows that the assumption of the PstI enzyme not working properly was wrong, as it shows the same result as the gel run on the 11/09/13.
Double digestion of the isolated pGEM plasmids ligated with the TOD operon genes (X, F and TOBG)
 "
"
