Exeter/25 July 2013
From 2013.igem.org
Fentwistle (Talk | contribs) |
|||
| (One intermediate revision not shown) | |||
| Line 9: | Line 9: | ||
We re-transformed: | We re-transformed: | ||
| - | *OmpR promoter (a.k.a | + | *OmpR promoter (a.k.a ''ompC''), BBa_R0082, AMP |
*CcaR green light sensor, BBa_K592002, CAM | *CcaR green light sensor, BBa_K592002, CAM | ||
*FixJ promoter (a.k.a FixL), BBa_K592006, CAM | *FixJ promoter (a.k.a FixL), BBa_K592006, CAM | ||
| Line 25: | Line 25: | ||
==Retry of cph8 gel== | ==Retry of cph8 gel== | ||
| - | We used two replicates of BBa_K322124 (cph8, our red light sensor), cph8 #4 and cph8 #7, which were correctly cut with various combinations of EcoRI, XbaI, SpeI and PstI, as shown below: | + | We used two replicates of BBa_K322124 (cph8, our red light sensor), cph8 #4 and cph8 #7, which were correctly cut with various combinations of <i>EcoRI</i>, <i>XbaI</i>, <i>SpeI</i> and <i>PstI</i>, as shown below: |
{| class="wikitable" | {| class="wikitable" | ||
| Line 37: | Line 37: | ||
| 3 || Control (no enzymes) #7 | | 3 || Control (no enzymes) #7 | ||
|- | |- | ||
| - | | 4 || EcoRI and PstI #4 | + | | 4 || <i>EcoRI</i> and <i>PstI</i> #4 |
|- | |- | ||
| - | | 5 || EcoRI and PstI #7 | + | | 5 || <i>EcoRI</i> and <i>PstI</i> #7 |
|- | |- | ||
| - | | 6 || EcoRI and SpeI #4 | + | | 6 || <i>EcoRI</i> and <i>SpeI</i> #4 |
|- | |- | ||
| - | | 7 || EcoRI and SpeI #7 | + | | 7 || <i>EcoRI</i> and <i>SpeI</i> #7 |
|- | |- | ||
| - | | 8 || PstI #4 | + | | 8 || <i>PstI</i> #4 |
|- | |- | ||
| - | | 9 || PstI #4 | + | | 9 || <i>PstI</i> #4 |
|- | |- | ||
| - | | 10 || PstI #7 | + | | 10 || <i>PstI</i> #7 |
|- | |- | ||
| - | | 11 || SpeI #4 | + | | 11 || <i>SpeI</i> #4 |
|- | |- | ||
| - | | 12 || SpeI #7 | + | | 12 || <i>SpeI</i> #7 |
|- | |- | ||
| - | | 13 || XbaI #4 | + | | 13 || <i>XbaI</i> #4 |
|- | |- | ||
| - | | 14 || XbaI #7 | + | | 14 || <i>XbaI</i> #7 |
|- | |- | ||
| - | | 15 || EcoRI #7 | + | | 15 || <i>EcoRI</i> #7 |
|- | |- | ||
| - | | 16 || EcoRI #4 | + | | 16 || <i>EcoRI</i> #4 |
|- | |- | ||
| 17 || 100bp Plus DNA Ladder | | 17 || 100bp Plus DNA Ladder | ||
|} | |} | ||
| - | (There are two replicates of PstI #4, as the first looked very pale and we were unsure about how much sample was present due to lack of loading dye. EcoRI #4 and EcoRI #7 were accidentally loaded in the wrong order. We used two different DNA ladders.) | + | (There are two replicates of <i>PstI</i> #4, as the first looked very pale and we were unsure about how much sample was present due to lack of loading dye. ,i>EcoRI</i> #4 and <i>EcoRI</i> #7 were accidentally loaded in the wrong order. We used two different DNA ladders.) |
[[Image:25th_july_cph8_gel.jpg|center||500px|Image: 500 pixels]] | [[Image:25th_july_cph8_gel.jpg|center||500px|Image: 500 pixels]] | ||
| - | As you can see from the gel, XbaI appears to have cut very oddly. The large "smear" of DNA is not expected, so we ran a gel which is just checking the ability of XbaI to cut cph8. | + | As you can see from the gel, <i>XbaI</i> appears to have cut very oddly. The large "smear" of DNA is not expected, so we ran a gel which is just checking the ability of <i>XbaI</i> to cut cph8. |
| - | We used two different vials of XbaI; one was kindly lent to us by another student in the lab. The iGEM XbaI will be referred to as XbaI (A), the borrowed XbaI is XbaI (B). The cph8 plasmids are cph8 #3 and cph8 #5. The control is cph8 cut with EcoRI. | + | We used two different vials of XbaI; one was kindly lent to us by another student in the lab. The iGEM <i>XbaI</i> will be referred to as <i>XbaI</i> (A), the borrowed <i>XbaI</i> is <i>XbaI</i> (B). The cph8 plasmids are cph8 #3 and cph8 #5. The control is cph8 cut with <i>EcoRI</i>. |
{| class="wikitable" | {| class="wikitable" | ||
| Line 80: | Line 80: | ||
| 1 || Ladder (1kb Gene Ruler) | | 1 || Ladder (1kb Gene Ruler) | ||
|- | |- | ||
| - | | 2 || XbaI (A) and cph8 #3 | + | | 2 || <i>XbaI</i> (A) and cph8 #3 |
|- | |- | ||
| - | | 3 || XbaI (A0 and cph8 #5 | + | | 3 || <i>XbaI</i> (A0 and cph8 #5 |
|- | |- | ||
| - | | 4 || XbaI (B) and cph8 #3 | + | | 4 || <i>XbaI</i> (B) and cph8 #3 |
|- | |- | ||
| - | | 5 || XbaI (B) and cph8 #5 | + | | 5 || <i>XbaI</i> (B) and cph8 #5 |
|- | |- | ||
| - | | 6 || EcoRI and cph8 #3 | + | | 6 || <i>EcoRI</i> and cph8 #3 |
|- | |- | ||
| - | | 7 || EcoRI and cph8 #5 | + | | 7 || <i>EcoRI</i> and cph8 #5 |
|- | |- | ||
| 8 || Ladder (1kb Gene Ruler) | | 8 || Ladder (1kb Gene Ruler) | ||
|} | |} | ||
| - | The gel showed that the cph8 plasmids were cut cleanly and uniformly. The previous gel, where XbaI appeared to not have worked, was probably due to human or technical error. It is important to remember that, during a digestion, the DNA in the Eppendorf has a habit of settling to the bottom of the tube, creating a minuscule and often unnoticed "pellet". You have to pipette from the bottom of the Eppendorf when loading the digestion into the gel, else there may be very little DNA present in the well. | + | The gel showed that the cph8 plasmids were cut cleanly and uniformly. The previous gel, where <i>XbaI</i> appeared to not have worked, was probably due to human or technical error. It is important to remember that, during a digestion, the DNA in the Eppendorf has a habit of settling to the bottom of the tube, creating a minuscule and often unnoticed "pellet". You have to pipette from the bottom of the Eppendorf when loading the digestion into the gel, else there may be very little DNA present in the well. |
Take me back to the [https://2013.igem.org/Team:Exeter/Notebook notebook]. | Take me back to the [https://2013.igem.org/Team:Exeter/Notebook notebook]. | ||
Latest revision as of 20:34, 2 October 2013
Transformations
We have decided to re-transform some of our BioBricks from the Kit Plates to make some reliable colonies and glycerol stocks.
We re-transformed:
- OmpR promoter (a.k.a ompC), BBa_R0082, AMP
- CcaR green light sensor, BBa_K592002, CAM
- FixJ promoter (a.k.a FixL), BBa_K592006, CAM
- YF1 blue light sensor, BBa_K592004, CAM
- RBS + cph8, BBa_K592018, CAM
- Terminator, BBa_B0015, CAM
- lambda inverter system, BBa_Q04510, KAN
- OmpR, BBa_K098011, CAM
- Promoter + RBS, BBa_K608002, CAM
This also includes re-plating the LB stabs we received from iGEM, which include Fix J (BBa_K592005), CcaS (BBa_K592001), Magenta (BBa_K592012), Yellow (BBa_K592010) and phycocyanobilin (codes for heme oxygenase and ferredoxin oxidoreducatse, BBa_K322122).
We also made streak plates of cph8 from our glycerol stocks (4 replicate plates).
Retry of cph8 gel
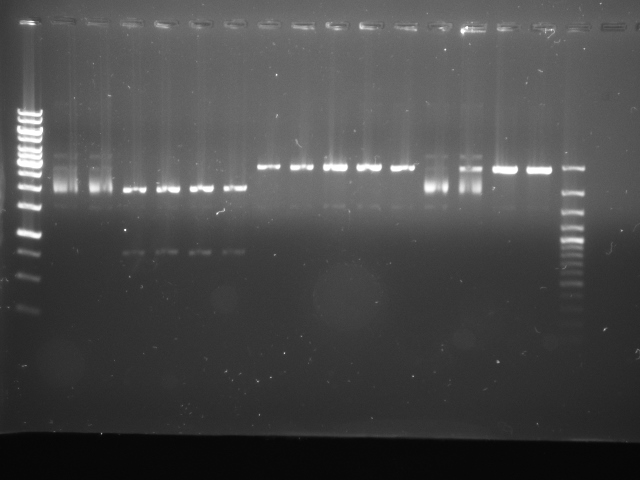
We used two replicates of BBa_K322124 (cph8, our red light sensor), cph8 #4 and cph8 #7, which were correctly cut with various combinations of EcoRI, XbaI, SpeI and PstI, as shown below:
| Lane | Contents |
|---|---|
| 1 | Ladder (1kb Gene Ruler) |
| 2 | Control (no enzymes) #4 |
| 3 | Control (no enzymes) #7 |
| 4 | EcoRI and PstI #4 |
| 5 | EcoRI and PstI #7 |
| 6 | EcoRI and SpeI #4 |
| 7 | EcoRI and SpeI #7 |
| 8 | PstI #4 |
| 9 | PstI #4 |
| 10 | PstI #7 |
| 11 | SpeI #4 |
| 12 | SpeI #7 |
| 13 | XbaI #4 |
| 14 | XbaI #7 |
| 15 | EcoRI #7 |
| 16 | EcoRI #4 |
| 17 | 100bp Plus DNA Ladder |
(There are two replicates of PstI #4, as the first looked very pale and we were unsure about how much sample was present due to lack of loading dye. ,i>EcoRI</i> #4 and EcoRI #7 were accidentally loaded in the wrong order. We used two different DNA ladders.)
As you can see from the gel, XbaI appears to have cut very oddly. The large "smear" of DNA is not expected, so we ran a gel which is just checking the ability of XbaI to cut cph8.
We used two different vials of XbaI; one was kindly lent to us by another student in the lab. The iGEM XbaI will be referred to as XbaI (A), the borrowed XbaI is XbaI (B). The cph8 plasmids are cph8 #3 and cph8 #5. The control is cph8 cut with EcoRI.
| Lane | Contents |
|---|---|
| 1 | Ladder (1kb Gene Ruler) |
| 2 | XbaI (A) and cph8 #3 |
| 3 | XbaI (A0 and cph8 #5 |
| 4 | XbaI (B) and cph8 #3 |
| 5 | XbaI (B) and cph8 #5 |
| 6 | EcoRI and cph8 #3 |
| 7 | EcoRI and cph8 #5 |
| 8 | Ladder (1kb Gene Ruler) |
The gel showed that the cph8 plasmids were cut cleanly and uniformly. The previous gel, where XbaI appeared to not have worked, was probably due to human or technical error. It is important to remember that, during a digestion, the DNA in the Eppendorf has a habit of settling to the bottom of the tube, creating a minuscule and often unnoticed "pellet". You have to pipette from the bottom of the Eppendorf when loading the digestion into the gel, else there may be very little DNA present in the well.
Take me back to the notebook.
 "
"