Friday, 28 Jun 2013
From 2013.igem.org
(Difference between revisions)
(Created page with "=='''6/28/13 Researching the Sequence of kerA'''== *Planning subteams continued to order reagents and compile protocols. *The plasmid design team decided on gibson assembly to p...") |
|||
| Line 4: | Line 4: | ||
*The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT. | *The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT. | ||
*kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995). | *kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995). | ||
| - | + | ||
| - | + | [[File:UChicago 2013 kerA plasmid design.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
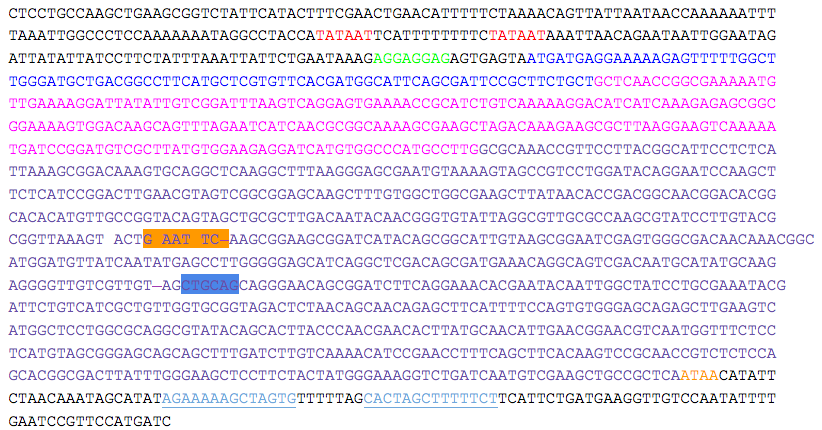
*Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated. | *Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated. | ||
*EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick. | *EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick. | ||
Revision as of 07:17, 26 September 2013
6/28/13 Researching the Sequence of kerA
- Planning subteams continued to order reagents and compile protocols.
- The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT.
- kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995).
- Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated.
- EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick.
 "
"