Team:Biwako Nagahama/Material & Method
From 2013.igem.org
IGEMBiwako (Talk | contribs) (Created page with "{{Biwako_Nagahama/header}} <div class="main-contents"> <div class="top-wrapper"> <!-- ここにtextを書いてください --> <div class="spacer"></div> <div class="top-...") |
|||
| (173 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<div class="spacer"></div> | <div class="spacer"></div> | ||
<div class="top-sentence"> | <div class="top-sentence"> | ||
| + | = <h2>Material & Method</h2> = | ||
| + | == <html> | ||
| + | <ul> | ||
| + | <li><a href="https://2013.igem.org/Team:Biwako_Nagahama/general protocol"><h2>General protocol</h2></a></li> | ||
| + | <ul> | ||
| - | |||
| - | |||
| - | + | </html> == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | == <h2>CelC</h2> == | ||
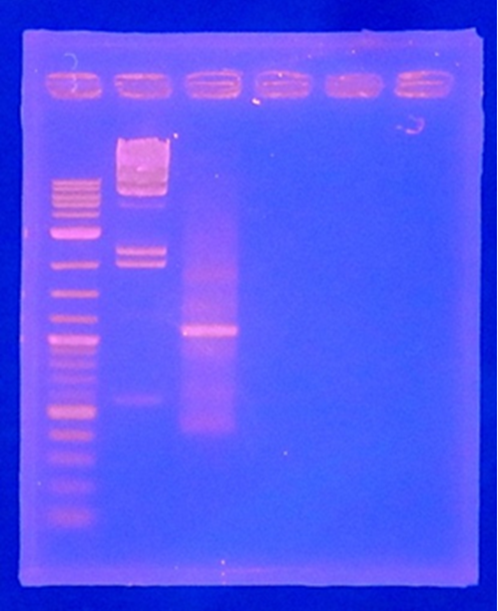
<P></p> | <P></p> | ||
<p>Agro Notebook</p> | <p>Agro Notebook</p> | ||
| Line 114: | Line 23: | ||
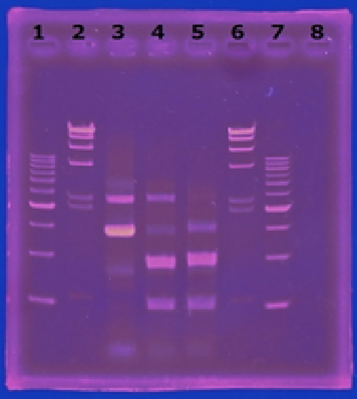
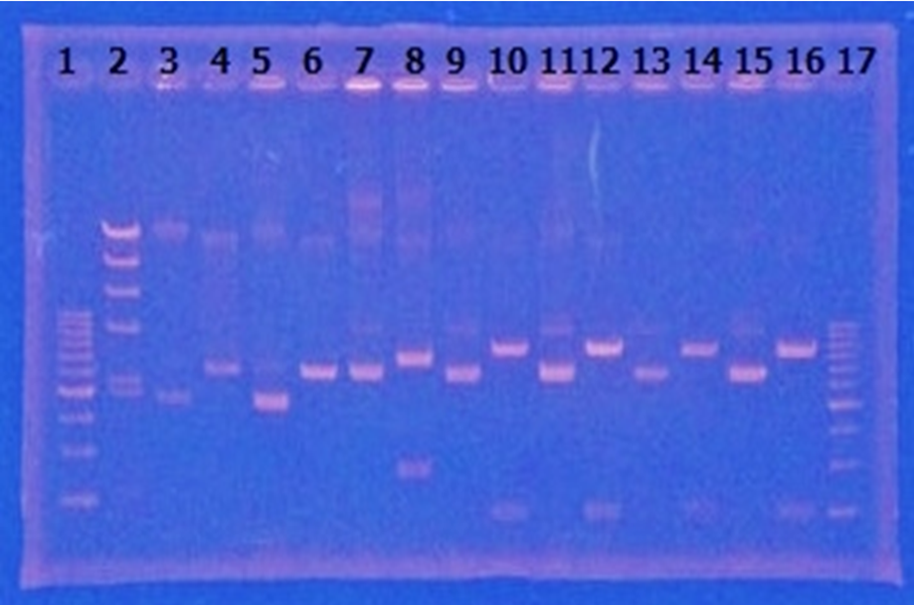
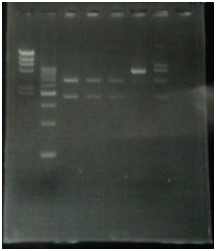
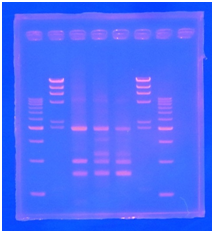
<p>CelC gene had produced clone from Agrobacterium tumefaciens C58, but I confirmed whether it’s true or not. CelC gene has restriction enzyme sites,EcoRI and BamHI. </p> | <p>CelC gene had produced clone from Agrobacterium tumefaciens C58, but I confirmed whether it’s true or not. CelC gene has restriction enzyme sites,EcoRI and BamHI. </p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
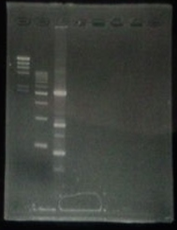
| - | + | [[File:Biwako-Nagahama_IGEM_訂正版図1.png|200px|left]] | |
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CelC_nonRE</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CelC/BamHI</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CelC/EcoRI</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>7</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
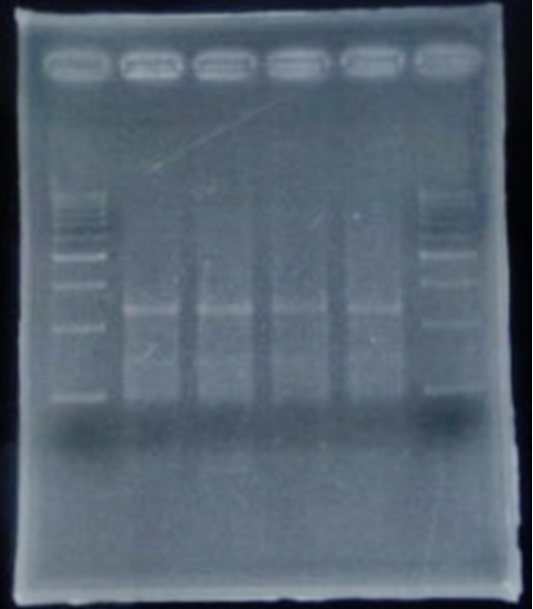
| + | <h3>Inverse PCR</h3> | ||
| + | <div style> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | |||
| + | <p> Inverse PCR | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>10xEX Taq Buffer </td><td>5μL</td></tr> | ||
| + | <tr><td>dNTP Mixture(2.5mM)</td><td>4μL</td></tr> | ||
| + | <tr><td>10pmol/µl Primer F</td><td>2.5μL</td></tr> | ||
| + | <tr><td>10pmol/µl Primer R</td><td>2.5μL</td></tr> | ||
| + | <tr><td>PCR反応液 No.9</td><td>1µl</td></tr> | ||
| + | <tr><td>Ex Taq HS (5U/µl)</td><td>0.25µl</td></tr> | ||
| + | <tr><td>dH2O </td><td>34.75µl</td></tr> | ||
| + | <tr><td>Total</td><td>50µl</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
</div> | </div> | ||
| + | <h3>Inverse PCR</h3> | ||
| + | <div style> | ||
| - | <div | + | |
| + | |||
| + | |||
| + | <p>※Restriction Enzyme sol.(line No.5 CelC_EcoRI)which has 1M NaCl dissolved in it, so, the band in lane no.5 appeared as upper-side. | ||
| + | </p> | ||
| + | <p> CelC_Fw: TGACGAAAGCACTGATCTGC | ||
| + | |||
| + | </p> | ||
| + | <p> CelC_Rv: GAAAAGATCGAAACGGTGG | ||
| + | </p> | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>94℃ 2min</td><td></td></tr> | ||
| + | <tr><td>↓</td><td></td></tr> | ||
| + | <tr><td></td><td></td></tr> | ||
| + | <tr><td>98℃ 10sec</td><td></td></tr> | ||
| + | <tr><td>49℃ 30sec</td><td>35cycles</td> | ||
| + | <tr><td>72℃ 2min</td><td></td></tr> | ||
| + | <tr><td> ↓</td><td></td></tr> | ||
| + | <tr><td>72℃ 2min</td><td></td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
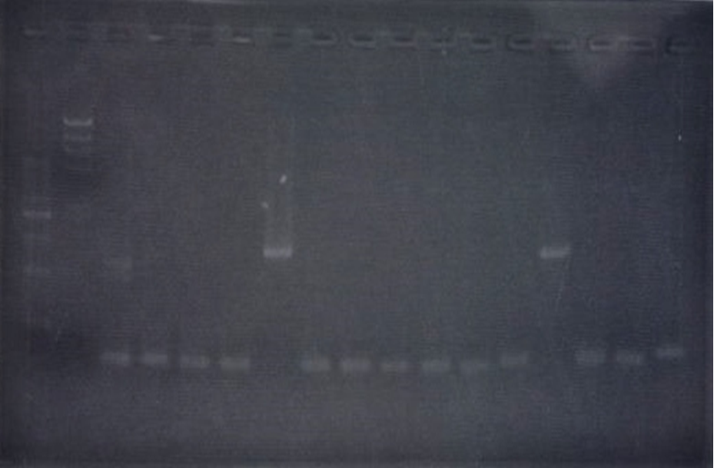
| + | <h3>Inverse PCR</h3> | ||
| + | <div style> | ||
| + | |||
| + | |||
| + | |||
| + | <p> TA cloning of CelC | ||
| + | </p> | ||
| + | |||
| + | [[File:Biwako-Nagahama T.Ksenpai celC4.png|500px|]] | ||
| + | |||
| + | <p>16℃ 30min incubate | ||
| + | </p> | ||
| + | <p> Ligation of CelC/pMD20 and Transformation in JM109. | ||
| + | </p> | ||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC5.png|500px|]] | ||
| + | |||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC6.png|500px|]] | ||
| + | |||
| + | <p> Cells were stored on ice for 30min. | ||
| + | </p> | ||
| + | <p> After 42℃ 30sec heat shock, cells were stored on ice for 2min. | ||
| + | </p> | ||
| + | <p> Then cells were pre-cultured at 37℃ for 1hr, plated to Ampicillin plate. | ||
| + | </p> | ||
| + | <p>6/1 Liquid clluture | ||
| + | </p> | ||
| + | <p> CelC/pMD20 22 samples at 37°C, for overnight. | ||
| + | </p> | ||
| + | <p>6/2 MiniPrep of CelC | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>2</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>4</td><td>Blue1</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>Blue2</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>Blue3</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>Blue4</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>Blue5</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>Blue6</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>White1</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>White2</td><td>12μL</td></tr> | ||
| + | <tr><td>12</td><td>White3</td><td>12μL</td></tr> | ||
| + | <tr><td>13</td><td>White4</td><td>12μL</td></tr> | ||
| + | <tr><td>14</td><td>White5</td><td>12μL</td></tr> | ||
| + | <tr><td>15</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>16</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>17</td><td>-</td><td>-</td></tr></p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>2</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>4</td><td>White6</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>White7</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>White8</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>White9</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>White10</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>White11</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>White12</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>White13</td><td>12μL</td></tr> | ||
| + | <tr><td>12</td><td>White14</td><td>12μL</td></tr> | ||
| + | <tr><td>13</td><td>White15</td><td>12μL</td></tr> | ||
| + | <tr><td>14</td><td>White16</td><td>12μL</td></tr> | ||
| + | <tr><td>15</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>16</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>17</td><td>500bp DNA ladder</td><td>10μL</td></tr></p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
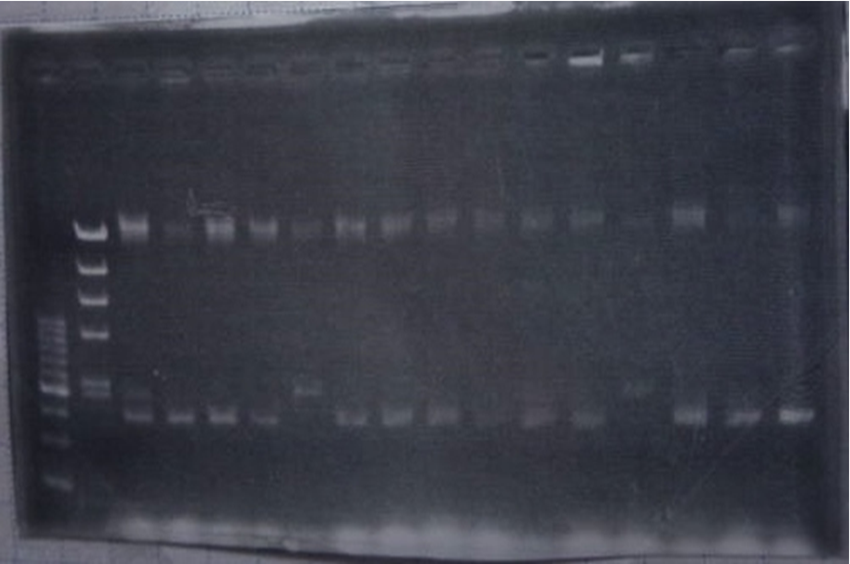
| + | <h3>Inverse PCR</h3> | ||
| + | <div style> | ||
| + | |||
| + | |||
| + | |||
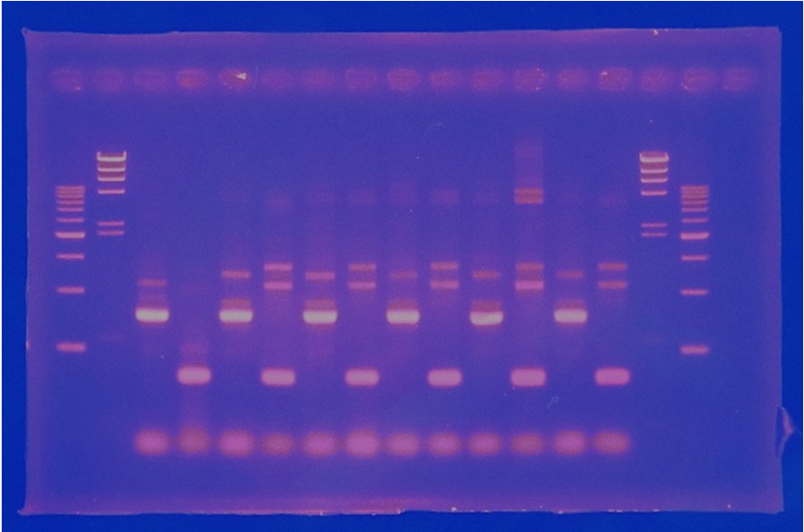
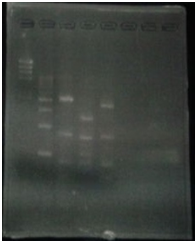
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC8.png|500px|]] | ||
| + | |||
| + | <p> Linear CelC/pMD20 DNA :2736bp. This CelC/pMD20 sample is cccDNA. So,white 6,white 7,white 8,white 9,white 14 probably picked up CelC/pMD20. | ||
| + | </p> | ||
| + | <p>6/3 Restriction Enzyme of CelC/pMD20 | ||
| + | </p> | ||
| + | <p> CelC gene had produced clone from Agrobacterium tumefaciens C58, I confirmed whether it’s true or not. CelC/pMD20 gene has 2 restriction enzyme sites,BamHI | ||
| + | </p> | ||
| + | |||
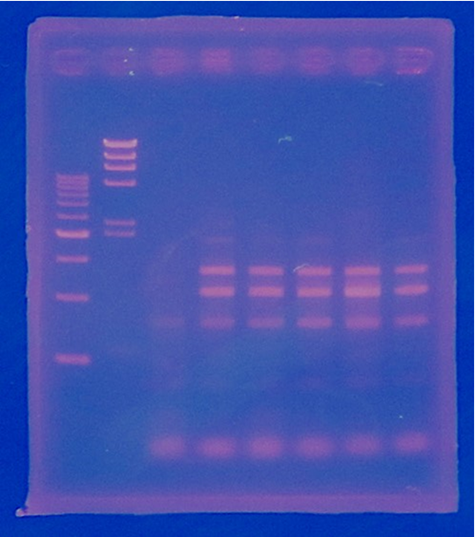
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC9.png|500px|]] | ||
| + | |||
| + | |||
| + | [[File:Biwako-Nagahama T.Ksenpai celC10.png|500px|]] | ||
| + | |||
| + | <p> I confirmed the direction of CelC gene. | ||
| + | </p> | ||
| + | <p>6/ Sequence of CelC/pMD20 | ||
| + | |||
| + | </p> | ||
| + | |||
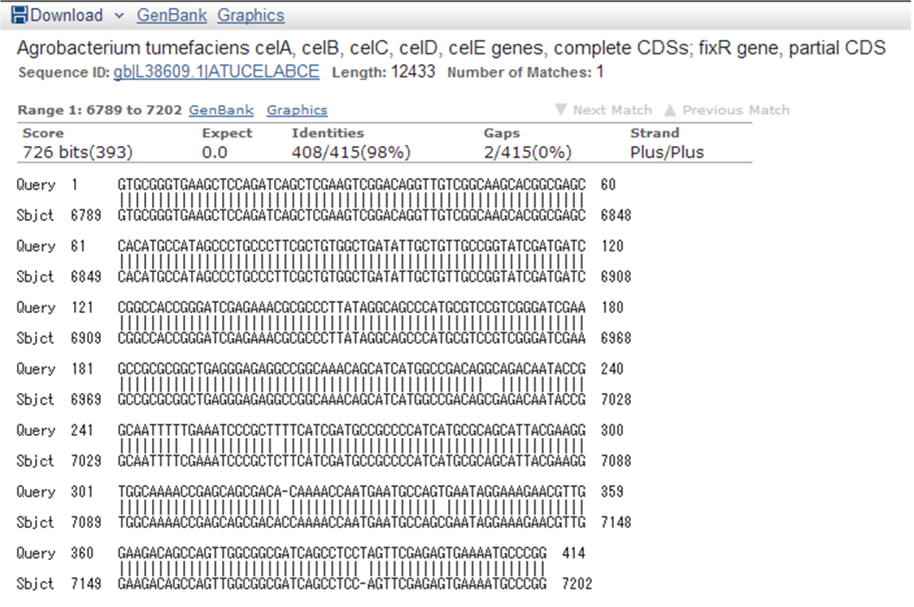
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC11.png|500px|]] | ||
| + | |||
| + | <p>7/18 PointMutation of CelC | ||
| + | </p> | ||
| + | <p> CelC gene had Restriction Enzyme Site,EcoRI. We directed the EcoRI site. | ||
| + | </p> | ||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC12.png|400px|left]] | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CelC Negative Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CelC Negative Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CelC No.6 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>CelC No.6 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>CelC No.7 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>CelC No.7 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>CelC No.8 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>CelC No.8 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>CelC No.9 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>12</td><td>CelC No.9 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>13</td><td>CelC No.14 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>14</td><td>CelC No.14 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>15</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>16</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <p> Front | ||
| + | </p> | ||
| + | <p> Fw1 Primer: | ||
| + | </p> | ||
| + | <p> TATATATTCTAGATGAAGAGCGGGATTTCG | ||
| + | |||
| + | </p> | ||
| + | <p> Rv2 Primer: | ||
| + | </p> | ||
| + | <p> CATTATATCCGAACTCCGGCTG | ||
| + | </p> | ||
| + | <p> Rear | ||
| + | </p> | ||
| + | <p> Fw2 Primer: | ||
| + | </p> | ||
| + | <p> AGCCGGAGTTCGGATATAATGC | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <p> Rv1 Primer: | ||
| + | </p> | ||
| + | <p> CAGCACGAACTAGTATTATTATCATCGGC | ||
| + | </p> | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>5×PS buffer</td><td>10μL</td></tr> | ||
| + | <tr><td>dNTP Mixture(2.5mM)</td><td>4μL</td></tr> | ||
| + | <tr><td>10pmol/μL Primer Fw1 or Fw2</td><td>1μL</td></tr> | ||
| + | <tr><td>10pmol/μL Primer Rv1 or Rv2</td><td>1μL</td></tr> | ||
| + | <tr><td>Template</td><td>1μL</td></tr> | ||
| + | <tr><td>Prime STAR HS(5U/μL)</td><td>0.5μL</td></tr> | ||
| + | <tr><td>dH2O</td><td>32.5μL</td></tr> | ||
| + | <tr><td>Total</td><td>50μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <p>7/19 Gel Purification of CelC gene’s Front Fragment and Rear Fragment | ||
| + | </p> | ||
| + | <p> Front Fragment DNA and Rear Fragment DNA that 765bp and 347bp band performed Gel Purification by illustra GFX PCR Purification Kit. | ||
| + | </p> | ||
| + | <p>7/21 CelC gene’s Front Fragment and Rear Fragment Overlap PCR | ||
| + | |||
| + | </p> | ||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC16.png|200px|left]] | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CelC N Assembly DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CelC No.6 Assembly DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CelC No.7 Assembly DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>CelC No.8 Assembly DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>CelC No.9 Assembly DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>CelC No.14 Assembly DNA</td><td>12μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <p> No.6,7,8,9,14 each fragment Overlap PCR completed. | ||
| + | |||
| + | </p> | ||
| + | <p> I selected No.8. | ||
| + | |||
| + | </p> | ||
| + | <p>7/22 Gel Purification of CelC gene No.8 | ||
| + | |||
| + | </p> | ||
| + | <p> No.8 DNA that about 1ooo bp band performed Gel Purification by illustra GFX PCR Purification Kit. | ||
| + | |||
| + | </p> | ||
| + | <p>8/1 Adapter PCR of CelC gene No.8 (BioBrick Part) | ||
| + | |||
| + | </p> | ||
| + | |||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC18.png|200px|left]] | ||
| + | |||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>2-log DNA ladder</td><td>5μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>6μL</td></tr> | ||
| + | <tr><td>3</td><td>PCR sample</td><td>18μL</td></tr> | ||
| + | <tr><td>4</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>5</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>6</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>7</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | <p> Fw Primer:GTTTCTTCGAATTCGCGGCCGCTTCTAGATG | ||
| + | </p> | ||
| + | <p> Rv Primer:GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTATC | ||
| + | </p> | ||
| + | <p>8/11 BioBrick of CelC Restrict Enzyme ,EcoRI. | ||
| + | |||
| + | </p> | ||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC20.png|200px|left]] | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>PCR sample A non RE</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>PCR sample A EcoRⅠ RE</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>PCR sample B non RE</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>PCR sample B EcoRⅠ RE</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>8</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <p> I confirmed BioBrick of CelC non-Restriction Enzyme ,EcoRI. | ||
| + | </p> | ||
| + | <p>9/5 Brick Part of CelC and pSB1C3 Restrict Enzyme,EcoRI and PstI. | ||
| + | |||
| + | </p> | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>・CelC</td><td></td></tr> | ||
| + | <tr><td>S.D.W</td><td>10μL</td></tr> | ||
| + | <tr><td>EcoRⅠ</td><td>1μL</td></tr> | ||
| + | <tr><td>PstⅠ</td><td>1μL</td></tr> | ||
| + | <tr><td>PCR sample</td><td>6μL</td></tr> | ||
| + | <tr><td>10×H buffer</td><td>2μL</td></tr> | ||
| + | <tr><td>Total</td><td>20μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>・pSB1C3</td><td></td></tr> | ||
| + | <tr><td>S.D.W</td><td>10μL</td></tr> | ||
| + | <tr><td>EcoRⅠ</td><td>1μL</td></tr> | ||
| + | <tr><td>PstⅠ</td><td>1μL</td></tr> | ||
| + | <tr><td>pSB1C3</td><td>6μL</td></tr> | ||
| + | <tr><td>10×H buffer</td><td>2μL</td></tr> | ||
| + | <tr><td>Total</td><td>20μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <p>9/5 Ligation of CelC/pSB1C3 and Transformation in JM109. | ||
| + | |||
| + | </p> | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>CelC/EcoRⅠ,PstⅠ(100ng)</td><td>1μL</td></tr> | ||
| + | <tr><td>pSB1C3/EcoRⅠ,PstⅠ(50ng)</td><td>2μL</td></tr> | ||
| + | <tr><td>Ligation kit ver.2</td><td>3μL</td></tr> | ||
| + | <tr><td>Total</td><td>6μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <p>9/7 Colony PCR of CelC/pSB1C3 | ||
| + | |||
| + | </p> | ||
| + | |||
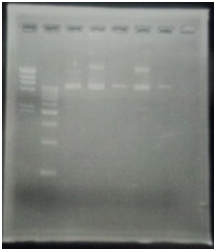
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC24.png|450px|left]] | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>No.1</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>No.2</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>No.3</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>No.4</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>No.5</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>No.6</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>No.7</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>No.8</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>No.9</td><td>12μL</td></tr> | ||
| + | <tr><td>12</td><td>No.10</td><td>12μL</td></tr> | ||
| + | <tr><td>13</td><td>No.11</td><td>12μL</td></tr> | ||
| + | <tr><td>14</td><td>No.12</td><td>12μL</td></tr> | ||
| + | <tr><td>15</td><td>No.13</td><td>12μL</td></tr> | ||
| + | <tr><td>16</td><td>No.14</td><td>12μL</td></tr> | ||
| + | <tr><td>17</td><td>No.15</td><td>12μL</td></tr> | ||
| + | <tr><td>18</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <p>CelC/pSB1C3 DNA(VR-VF2) :1370bp. So,No.5 and No.12 probably picked up CelC/pSB1C3 | ||
| + | </p> | ||
| + | <p>9/8 Miniprep of CelC/pSB1C3 | ||
| + | |||
| + | </p> | ||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC27.png|450px|left]] | ||
| + | |||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>No.1</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>No.2</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>No.3</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>No.4</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>No.5</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>No.6</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>No.7</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>No.8</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>No.9</td><td>12μL</td></tr> | ||
| + | <tr><td>12</td><td>No.10</td><td>12μL</td></tr> | ||
| + | <tr><td>13</td><td>No.11</td><td>12μL</td></tr> | ||
| + | <tr><td>14</td><td>No.12</td><td>12μL</td></tr> | ||
| + | <tr><td>15</td><td>No.13</td><td>12μL</td></tr> | ||
| + | <tr><td>16</td><td>No.14</td><td>12μL</td></tr> | ||
| + | <tr><td>17</td><td>No.15</td><td>12μL</td></tr> | ||
| + | <tr><td>18</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <p> Linear CelC/pSB1C3 DNA :3126bp. This CelC/pSB1C3 sample is cccDNA. So,No.5 and No.12 probably picked up CelC/pSB1C3. | ||
| + | |||
| + | |||
| + | </p> | ||
| + | <p>9/15. Sequence of CelC Brick Part sequence. | ||
| + | |||
| + | </p> | ||
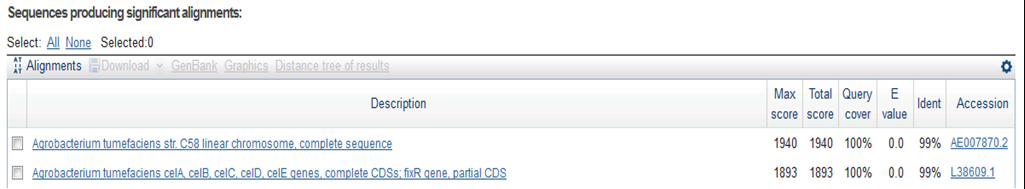
| + | <p> Result of NCBI BLAST | ||
| + | </p> | ||
| + | |||
| + | |||
| + | [[File:Biwako-Nagahama_T.Ksenpai_celC28.png|500px]] | ||
| + | |||
| + | |||
| + | === <h2> CrdS</h2> === | ||
| + | <h3>Cloning of CrdS and Restriction Enzyme</h3> | ||
| + | <h5>By Koki Tsutsumi</h5> | ||
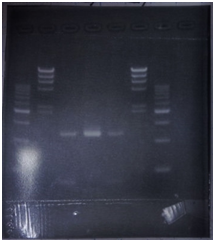
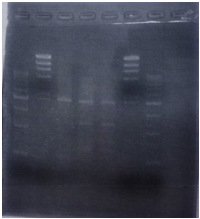
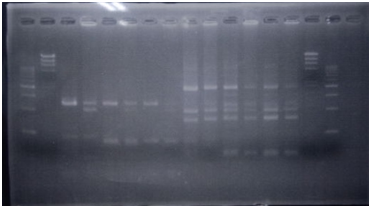
| + | <p>CrdS gene had produced clone from Agrobacterium tumefaciens C58, but I confirmed whether it’s true or not. CrdS gene has restriction enzyme sites,EcoRI and PstI. </p> | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi1.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>5</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>6</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>7</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi2.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS_nonRE</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS/EcoRⅠ</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS/PstⅠ</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>7</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <h3>Inverse PCR</h3> | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>Prime STAR MAX</td><td>25μL</td></tr> | ||
| + | <tr><td>10pmol/μL Primer F</td><td>1μL</td></tr> | ||
| + | <tr><td>10pmol/μL Primer R</td><td>1μL</td></tr> | ||
| + | <tr><td>PCR反応液(2ng)</td><td>1μL</td></tr> | ||
| + | <tr><td>dH2O</td><td>22μL</td></tr> | ||
| + | <tr><td>Total</td><td>22μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <p>CrdS_Fw: AGTACGATCCGCTATTTTCCCG</p> | ||
| + | <p>CrdS_Rv: CAGACCAAGATTTCGCGAACTC</p> | ||
| + | ---- | ||
| + | <p>94℃ 2min</p> | ||
| + | <p>↓</p> | ||
| + | <p>98℃ 10sec</p> | ||
| + | <p>48℃ 30sec 35cycles</p> | ||
| + | <p>72℃ 2min</p> | ||
| + | <p>↓</p> | ||
| + | <p>72℃ 2min</p> | ||
| + | ---- | ||
| + | <h3>TA cloning of CrdS</h3> | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <tr><td>PCR product</td><td>1μL</td></tr> | ||
| + | <tr><td>pMD20</td><td>2μL</td></tr> | ||
| + | <tr><td>Ligation kit ver.2</td><td>3μL</td></tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | 16℃ 30min incubate | ||
| + | <h3>Ligation of CrdS/pMD20 and Transformation in JM109</h3> | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <tr><td>Competent cell</td><td>100μL</td></tr> | ||
| + | <tr><td>DNA</td><td>6μL</td></tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | <p>↓Cells were stored on ice for 30min. </p> | ||
| + | <p>↓After 42℃ 30sec heat shock, cells were stored on ice for 2min.</p> | ||
| + | <p>↓Then cells were pre-cultured at 37℃ for 1hr, plated to Ampicillin plate. </p> | ||
| + | <h3>Liquid culluture</h3> | ||
| + | CrdS/pMD20 21 samples at 37℃,for overnight | ||
| + | <h3>MiniPrep of CrdS/pMD20</h3> | ||
| + | Linear CrdS/pMD20 DNA :4995bp. This CrdS/pMD20 sample is cccDNA. So, probably picked up CrdS/pMD20. | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi3.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS No.5</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS No.6</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS No.8</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>CrdS No.15</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>CrdS No.17</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <h3>Restriction Enzyme of CrdS/pMD20</h3> | ||
| + | CrdS gene had produced clone from Agrobacterium tumefaciens C58, I confirmed whether it’s true or not. CrdS/pMD20 gene has 2 restriction enzyme sites,EcoRI. | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi4.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS No.5</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS No.6</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS No.2</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>CrdS No.15</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>CrdS No.17</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | I comfirmed the direction of CrdS gene. | ||
| + | <h3>Sequence of CrdS/pMD20</h3> | ||
| + | [[File:Biwako-Nagahama_Sequences_tutumi5.png|700px|]] | ||
| + | <h3>PointMutation of CrdS</h3> | ||
| + | CrdS gene had Restriction Enzyme Site,EcoRI. We removed the EcoRI site. | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi6.png|left|]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS Negative Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS Negative Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS No.6 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>CrdS No.6 Middle Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>CrdS No.6 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>CrdS No.15 Front Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>CrdS No.15 Middle Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>CrdS No.15 Rear Fragment</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>12</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>13</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>14</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>15</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>16</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <div style='width:560;font:bold 14;color:black;text-align:left;> | ||
| + | <p>Front</p> | ||
| + | <p>Fw0 Primer:GGCCGCTTCTAGATGTATTTCAGTGC</p> | ||
| + | <p>Rv1 Primer:CGTTTCGAGGGAGAACTCCAGCG</p> | ||
| + | <p>Middle</p> | ||
| + | <p>Fw1Primer:CGCTGGAGTTCTCCCTCGAAACG</p> | ||
| + | <p>Rv2Primer:CAGCTCATCGGCCTGAAGCGCC</p> | ||
| + | <p>Rear</p> | ||
| + | <p>Fw2 Primer:GGCGCTTCAGGCCGATGAGCTG</p> | ||
| + | <p>Rv0 Primer:GGCGCTACTAGTATTATTATCACCCGAATG</p> | ||
| + | </div> | ||
| + | <div align="left"> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>5xPS Buffer</td><td>10μL</td></tr> | ||
| + | <tr><td>dNTP Mixture</td><td>4μL</td></tr> | ||
| + | <tr><td>10pmol/µl Primer Fw1 or Fw2</td><td>1μL</td></tr> | ||
| + | <tr><td>10pmol/µl Primer Rv2 or Rv1</td><td>1μL</td></tr> | ||
| + | <tr><td>Templete</td><td>1μL</td></tr> | ||
| + | <tr><td>Prime STAR HS</td><td>0.5μL</td></tr> | ||
| + | <tr><td>dH2O</td><td>32.5μL</td></tr> | ||
| + | <tr><td>Total</td><td>50μL</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | ---- | ||
| + | <p>94℃ 2min</p> | ||
| + | <p>↓</p> | ||
| + | <p>98℃ 10sec</p> | ||
| + | <p>55℃ 5sec 35cycles</p> | ||
| + | <p>72℃ 70sec</p> | ||
| + | <p>↓</p> | ||
| + | <p>4℃ ∞</p> | ||
| + | ---- | ||
| + | <h3>Gel Purification of CrdS gene’s Front Fragment and Rear Fragment</h3> | ||
| + | Front Fragment DNA and Rear Fragment DNA ,Middle Fragment DNA that 223 bp and 1037 bp ,782 bp band performed Gel Purification by illustra GFX PCR Purification Kit. | ||
| + | <h3>CrdS gene’s Front Fragment and Rear Fragment Overlap PCR</h3> | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi7.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS N Front+Middle DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS No.6 Front+Middle DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS No.15 Front+Middle DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>7</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi8.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS N Front+Rear DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS No.6 Front+Rear DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS No.15 Front+Rear DNA</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>7</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | N, No.6, No.15 each fragment Overlap PCR completed. | ||
| + | <h3>Gel Purification of CrdS</h3> | ||
| + | No.8 DNA that about 2ooo bp band performed Gel Purification by illustra GFX PCR Purification Kit. | ||
| + | <h3>Adapter PCR of CrdS gene (BioBrick Part)</h3> | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi9.png|left|170px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p> | ||
| + | <tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>2-log DNA ladder</td><td>5μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>6μL</td></tr> | ||
| + | <tr><td>3</td><td>PCR saample</td><td>18μL</td></tr> | ||
| + | <tr><td>4</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>5</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>6</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>7</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>8</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
| + | </div> | ||
| + | <p>Fw Primer: | ||
| + | GTTTCTTCGAATTCGCGGCCGCTTCTAGATG</p> | ||
| + | <p>Rv Primer: | ||
| + | GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTATC</p> | ||
| + | <h3>Brick Part of CrdS Restrict Enzyme,EcoRI and PstI.</h3> | ||
| + | [[File:Biwako-Nagahama_E.P_tutumi10.png|left|450px]] | ||
| + | <div style> | ||
| + | <table border="1"> | ||
| + | <p><tr><td>No</td><td>Name</td><td>volume</td></tr> | ||
| + | <tr><td>1</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>2</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>3</td><td>CrdS N F+M non-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>4</td><td>CrdS N F+M EcoRⅠ-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>5</td><td>CrdS No.6 F+M non-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>6</td><td>CrdS No.6 F+M EcoRⅠ-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>7</td><td>CrdS No.15 F+M non-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>8</td><td>CrdS No.15 F+M EcoRⅠ</td><td>12μL</td></tr> | ||
| + | <tr><td>9</td><td>CrdS N F+M+R non-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>10</td><td>CrdS N F+M+R PstⅠ-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>11</td><td>CrdS No.6 F+M+R non-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>12</td><td>Crds No.6 F+M+R PstI RE</td><td>12μL</td></tr> | ||
| + | <tr><td>13</td><td>CrdS No.15 F+M+R non-RE</td><td>12μL</td></tr> | ||
| + | <tr><td>14</td><td>CrdS No.15 F+M+R PstI RE</td><td>12μL</td></tr> | ||
| + | <tr><td>15</td><td>λ-HindⅢ</td><td>10μL</td></tr> | ||
| + | <tr><td>16</td><td>500bp DNA ladder</td><td>10μL</td></tr> | ||
| + | <tr><td>17</td><td>-</td><td>-</td></tr> | ||
| + | <tr><td>18</td><td>-</td><td>-</td></tr> | ||
| + | </p> | ||
| + | </table> | ||
</div> | </div> | ||
| + | N,No.6,No.15 of CrdS was cut on EcoRI site. Point Mutation of CrdS is unsuccessful. | ||
Latest revision as of 02:31, 28 September 2013
Material & Method
CelC
Agro Notebook
5/31 Cloning of CelC and Restriction Enzyme
By Koki Tsutsumi
CelC gene had produced clone from Agrobacterium tumefaciens C58, but I confirmed whether it’s true or not. CelC gene has restriction enzyme sites,EcoRI and BamHI.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CelC_nonRE | 12μL |
| 4 | CelC/BamHI | 12μL |
| 5 | CelC/EcoRI | 12μL |
| 6 | λ-HindⅢ | 10μL |
| 7 | 500bp DNA ladder | 10μL |
| 8 | - | - |
Inverse PCR
Inverse PCR
| Name | volume |
| 10xEX Taq Buffer | 5μL |
| dNTP Mixture(2.5mM) | 4μL |
| 10pmol/µl Primer F | 2.5μL |
| 10pmol/µl Primer R | 2.5μL |
| PCR反応液 No.9 | 1µl |
| Ex Taq HS (5U/µl) | 0.25µl |
| dH2O | 34.75µl |
| Total | 50µl |
Inverse PCR
※Restriction Enzyme sol.(line No.5 CelC_EcoRI)which has 1M NaCl dissolved in it, so, the band in lane no.5 appeared as upper-side.
CelC_Fw: TGACGAAAGCACTGATCTGC
CelC_Rv: GAAAAGATCGAAACGGTGG
| 94℃ 2min | |
| ↓ | |
| 98℃ 10sec | |
| 49℃ 30sec | 35cycles |
| 72℃ 2min | |
| ↓ | |
| 72℃ 2min |
Inverse PCR
TA cloning of CelC
16℃ 30min incubate
Ligation of CelC/pMD20 and Transformation in JM109.
Cells were stored on ice for 30min.
After 42℃ 30sec heat shock, cells were stored on ice for 2min.
Then cells were pre-cultured at 37℃ for 1hr, plated to Ampicillin plate.
6/1 Liquid clluture
CelC/pMD20 22 samples at 37°C, for overnight.
6/2 MiniPrep of CelC
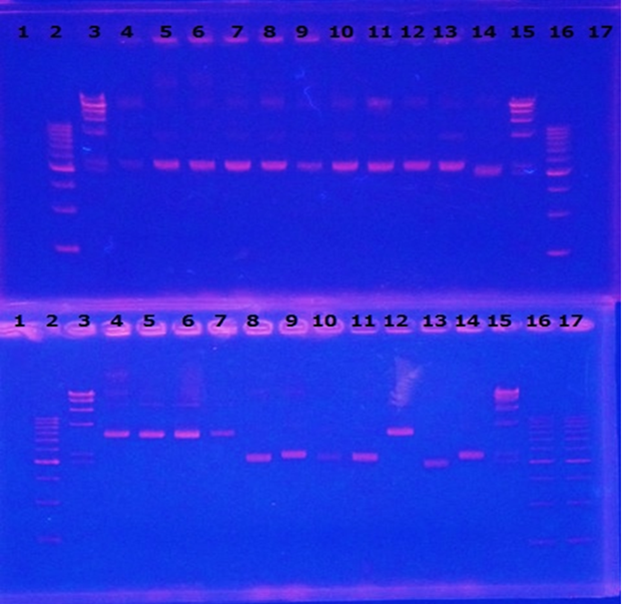
| No | Name | volume |
| 1 | - | - |
| 2 | 500bp DNA ladder | 10μL |
| 3 | λ-HindⅢ | 10μL |
| 4 | Blue1 | 12μL |
| 5 | Blue2 | 12μL |
| 6 | Blue3 | 12μL |
| 7 | Blue4 | 12μL |
| 8 | Blue5 | 12μL |
| 9 | Blue6 | 12μL |
| 10 | White1 | 12μL |
| 11 | White2 | 12μL |
| 12 | White3 | 12μL |
| 13 | White4 | 12μL |
| 14 | White5 | 12μL |
| 15 | λ-HindⅢ | 10μL |
| 16 | 500bp DNA ladder | 10μL |
| 17 | - | - |
| No | Name | volume |
| 1 | - | - |
| 2 | 500bp DNA ladder | 10μL |
| 3 | λ-HindⅢ | 10μL |
| 4 | White6 | 12μL |
| 5 | White7 | 12μL |
| 6 | White8 | 12μL |
| 7 | White9 | 12μL |
| 8 | White10 | 12μL |
| 9 | White11 | 12μL |
| 10 | White12 | 12μL |
| 11 | White13 | 12μL |
| 12 | White14 | 12μL |
| 13 | White15 | 12μL |
| 14 | White16 | 12μL |
| 15 | λ-HindⅢ | 10μL |
| 16 | 500bp DNA ladder | 10μL |
| 17 | 500bp DNA ladder | 10μL |
Inverse PCR
Linear CelC/pMD20 DNA :2736bp. This CelC/pMD20 sample is cccDNA. So,white 6,white 7,white 8,white 9,white 14 probably picked up CelC/pMD20.
6/3 Restriction Enzyme of CelC/pMD20
CelC gene had produced clone from Agrobacterium tumefaciens C58, I confirmed whether it’s true or not. CelC/pMD20 gene has 2 restriction enzyme sites,BamHI
I confirmed the direction of CelC gene.
6/ Sequence of CelC/pMD20
7/18 PointMutation of CelC
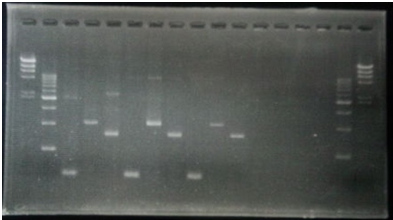
CelC gene had Restriction Enzyme Site,EcoRI. We directed the EcoRI site.
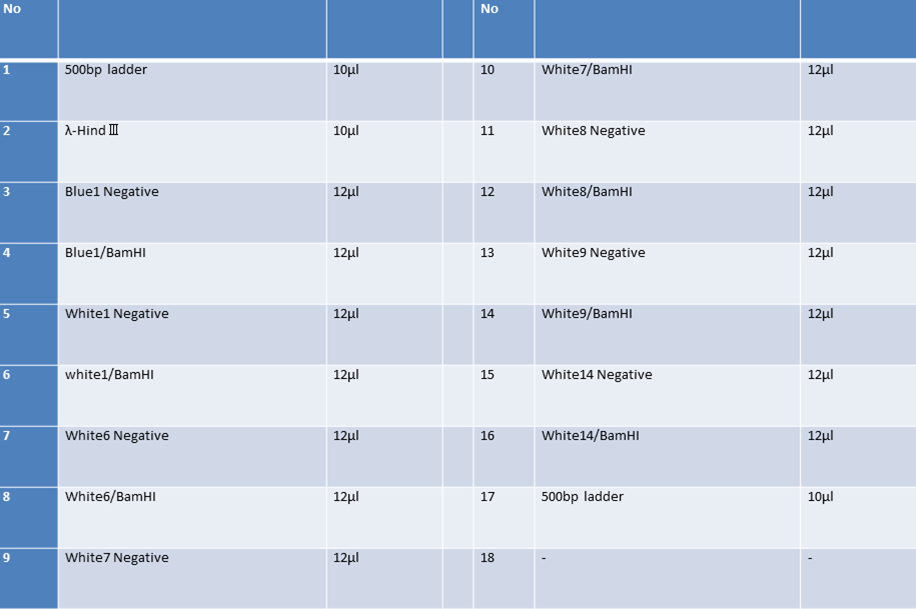
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CelC Negative Front Fragment | 12μL |
| 4 | CelC Negative Rear Fragment | 12μL |
| 5 | CelC No.6 Front Fragment | 12μL |
| 6 | CelC No.6 Rear Fragment | 12μL |
| 7 | CelC No.7 Front Fragment | 12μL |
| 8 | CelC No.7 Rear Fragment | 12μL |
| 9 | CelC No.8 Front Fragment | 12μL |
| 10 | CelC No.8 Rear Fragment | 12μL |
| 11 | CelC No.9 Front Fragment | 12μL |
| 12 | CelC No.9 Rear Fragment | 12μL |
| 13 | CelC No.14 Front Fragment | 12μL |
| 14 | CelC No.14 Rear Fragment | 12μL |
| 15 | λ-HindⅢ | 10μL |
| 16 | 500bp DNA ladder | 10μL |
Front
Fw1 Primer:
TATATATTCTAGATGAAGAGCGGGATTTCG
Rv2 Primer:
CATTATATCCGAACTCCGGCTG
Rear
Fw2 Primer:
AGCCGGAGTTCGGATATAATGC
Rv1 Primer:
CAGCACGAACTAGTATTATTATCATCGGC
| 5×PS buffer | 10μL |
| dNTP Mixture(2.5mM) | 4μL |
| 10pmol/μL Primer Fw1 or Fw2 | 1μL |
| 10pmol/μL Primer Rv1 or Rv2 | 1μL |
| Template | 1μL |
| Prime STAR HS(5U/μL) | 0.5μL |
| dH2O | 32.5μL |
| Total | 50μL |
7/19 Gel Purification of CelC gene’s Front Fragment and Rear Fragment
Front Fragment DNA and Rear Fragment DNA that 765bp and 347bp band performed Gel Purification by illustra GFX PCR Purification Kit.
7/21 CelC gene’s Front Fragment and Rear Fragment Overlap PCR
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CelC N Assembly DNA | 12μL |
| 4 | CelC No.6 Assembly DNA | 12μL |
| 5 | CelC No.7 Assembly DNA | 12μL |
| 6 | CelC No.8 Assembly DNA | 12μL |
| 7 | CelC No.9 Assembly DNA | 12μL |
| 8 | CelC No.14 Assembly DNA | 12μL |
No.6,7,8,9,14 each fragment Overlap PCR completed.
I selected No.8.
7/22 Gel Purification of CelC gene No.8
No.8 DNA that about 1ooo bp band performed Gel Purification by illustra GFX PCR Purification Kit.
8/1 Adapter PCR of CelC gene No.8 (BioBrick Part)
| No | Name | volume |
| 1 | 2-log DNA ladder | 5μL |
| 2 | λ-HindⅢ | 6μL |
| 3 | PCR sample | 18μL |
| 4 | - | - |
| 5 | - | - |
| 6 | - | - |
| 7 | - | - |
| 8 | - | - |
Fw Primer:GTTTCTTCGAATTCGCGGCCGCTTCTAGATG
Rv Primer:GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTATC
8/11 BioBrick of CelC Restrict Enzyme ,EcoRI.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | PCR sample A non RE | 12μL |
| 4 | PCR sample A EcoRⅠ RE | 12μL |
| 5 | PCR sample B non RE | 12μL |
| 6 | PCR sample B EcoRⅠ RE | 12μL |
| 7 | λ-HindⅢ | 10μL |
| 8 | 500bp DNA ladder | 10μL |
I confirmed BioBrick of CelC non-Restriction Enzyme ,EcoRI.
9/5 Brick Part of CelC and pSB1C3 Restrict Enzyme,EcoRI and PstI.
| ・CelC | |
| S.D.W | 10μL |
| EcoRⅠ | 1μL |
| PstⅠ | 1μL |
| PCR sample | 6μL |
| 10×H buffer | 2μL |
| Total | 20μL |
| ・pSB1C3 | |
| S.D.W | 10μL |
| EcoRⅠ | 1μL |
| PstⅠ | 1μL |
| pSB1C3 | 6μL |
| 10×H buffer | 2μL |
| Total | 20μL |
9/5 Ligation of CelC/pSB1C3 and Transformation in JM109.
| CelC/EcoRⅠ,PstⅠ(100ng) | 1μL |
| pSB1C3/EcoRⅠ,PstⅠ(50ng) | 2μL |
| Ligation kit ver.2 | 3μL |
| Total | 6μL |
9/7 Colony PCR of CelC/pSB1C3
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | No.1 | 12μL |
| 4 | No.2 | 12μL |
| 5 | No.3 | 12μL |
| 6 | No.4 | 12μL |
| 7 | No.5 | 12μL |
| 8 | No.6 | 12μL |
| 9 | No.7 | 12μL |
| 10 | No.8 | 12μL |
| 11 | No.9 | 12μL |
| 12 | No.10 | 12μL |
| 13 | No.11 | 12μL |
| 14 | No.12 | 12μL |
| 15 | No.13 | 12μL |
| 16 | No.14 | 12μL |
| 17 | No.15 | 12μL |
| 18 | - | - |
CelC/pSB1C3 DNA(VR-VF2) :1370bp. So,No.5 and No.12 probably picked up CelC/pSB1C3
9/8 Miniprep of CelC/pSB1C3
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | No.1 | 12μL |
| 4 | No.2 | 12μL |
| 5 | No.3 | 12μL |
| 6 | No.4 | 12μL |
| 7 | No.5 | 12μL |
| 8 | No.6 | 12μL |
| 9 | No.7 | 12μL |
| 10 | No.8 | 12μL |
| 11 | No.9 | 12μL |
| 12 | No.10 | 12μL |
| 13 | No.11 | 12μL |
| 14 | No.12 | 12μL |
| 15 | No.13 | 12μL |
| 16 | No.14 | 12μL |
| 17 | No.15 | 12μL |
| 18 | - | - |
Linear CelC/pSB1C3 DNA :3126bp. This CelC/pSB1C3 sample is cccDNA. So,No.5 and No.12 probably picked up CelC/pSB1C3.
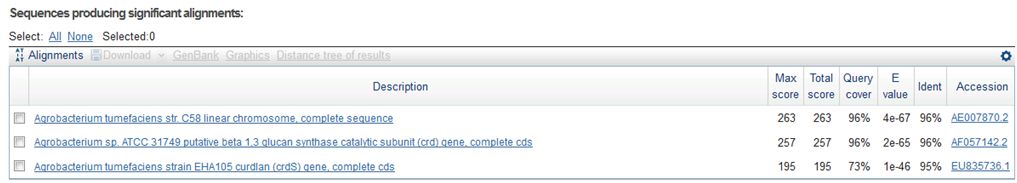
9/15. Sequence of CelC Brick Part sequence.
Result of NCBI BLAST
CrdS
Cloning of CrdS and Restriction Enzyme
By Koki Tsutsumi
CrdS gene had produced clone from Agrobacterium tumefaciens C58, but I confirmed whether it’s true or not. CrdS gene has restriction enzyme sites,EcoRI and PstI.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS | 12μL |
| 4 | - | - |
| 5 | - | - |
| 6 | - | - |
| 7 | - | - |
| 8 | - | - |
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS_nonRE | 12μL |
| 4 | CrdS/EcoRⅠ | 12μL |
| 5 | CrdS/PstⅠ | 12μL |
| 6 | λ-HindⅢ | 10μL |
| 7 | 500bp DNA ladder | 10μL |
| 8 | - | - |
Inverse PCR
| Prime STAR MAX | 25μL |
| 10pmol/μL Primer F | 1μL |
| 10pmol/μL Primer R | 1μL |
| PCR反応液(2ng) | 1μL |
| dH2O | 22μL |
| Total | 22μL |
CrdS_Fw: AGTACGATCCGCTATTTTCCCG
CrdS_Rv: CAGACCAAGATTTCGCGAACTC
94℃ 2min
↓
98℃ 10sec
48℃ 30sec 35cycles
72℃ 2min
↓
72℃ 2min
TA cloning of CrdS
| PCR product | 1μL |
| pMD20 | 2μL |
| Ligation kit ver.2 | 3μL |
16℃ 30min incubate
Ligation of CrdS/pMD20 and Transformation in JM109
| Competent cell | 100μL |
| DNA | 6μL |
↓Cells were stored on ice for 30min.
↓After 42℃ 30sec heat shock, cells were stored on ice for 2min.
↓Then cells were pre-cultured at 37℃ for 1hr, plated to Ampicillin plate.
Liquid culluture
CrdS/pMD20 21 samples at 37℃,for overnight
MiniPrep of CrdS/pMD20
Linear CrdS/pMD20 DNA :4995bp. This CrdS/pMD20 sample is cccDNA. So, probably picked up CrdS/pMD20.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS No.5 | 12μL |
| 4 | CrdS No.6 | 12μL |
| 5 | CrdS No.8 | 12μL |
| 6 | CrdS No.15 | 12μL |
| 7 | CrdS No.17 | 12μL |
| 8 | - | - |
Restriction Enzyme of CrdS/pMD20
CrdS gene had produced clone from Agrobacterium tumefaciens C58, I confirmed whether it’s true or not. CrdS/pMD20 gene has 2 restriction enzyme sites,EcoRI.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS No.5 | 12μL |
| 4 | CrdS No.6 | 12μL |
| 5 | CrdS No.2 | 12μL |
| 6 | CrdS No.15 | 12μL |
| 7 | CrdS No.17 | 12μL |
| 8 | - | - |
I comfirmed the direction of CrdS gene.
Sequence of CrdS/pMD20
PointMutation of CrdS
CrdS gene had Restriction Enzyme Site,EcoRI. We removed the EcoRI site.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS Negative Front Fragment | 12μL |
| 4 | CrdS Negative Rear Fragment | 12μL |
| 5 | CrdS No.6 Front Fragment | 12μL |
| 6 | CrdS No.6 Middle Fragment | 12μL |
| 7 | CrdS No.6 Rear Fragment | 12μL |
| 8 | CrdS No.15 Front Fragment | 12μL |
| 9 | CrdS No.15 Middle Fragment | 12μL |
| 10 | CrdS No.15 Rear Fragment | 12μL |
| 11 | - | - |
| 12 | - | - |
| 13 | - | - |
| 14 | - | - |
| 15 | λ-HindⅢ | 10μL |
| 16 | 500bp DNA ladder | 10μL |
Front
Fw0 Primer:GGCCGCTTCTAGATGTATTTCAGTGC
Rv1 Primer:CGTTTCGAGGGAGAACTCCAGCG
Middle
Fw1Primer:CGCTGGAGTTCTCCCTCGAAACG
Rv2Primer:CAGCTCATCGGCCTGAAGCGCC
Rear
Fw2 Primer:GGCGCTTCAGGCCGATGAGCTG
Rv0 Primer:GGCGCTACTAGTATTATTATCACCCGAATG
| 5xPS Buffer | 10μL |
| dNTP Mixture | 4μL |
| 10pmol/µl Primer Fw1 or Fw2 | 1μL |
| 10pmol/µl Primer Rv2 or Rv1 | 1μL |
| Templete | 1μL |
| Prime STAR HS | 0.5μL |
| dH2O | 32.5μL |
| Total | 50μL |
94℃ 2min
↓
98℃ 10sec
55℃ 5sec 35cycles
72℃ 70sec
↓
4℃ ∞
Gel Purification of CrdS gene’s Front Fragment and Rear Fragment
Front Fragment DNA and Rear Fragment DNA ,Middle Fragment DNA that 223 bp and 1037 bp ,782 bp band performed Gel Purification by illustra GFX PCR Purification Kit.
CrdS gene’s Front Fragment and Rear Fragment Overlap PCR
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS N Front+Middle DNA | 12μL |
| 4 | CrdS No.6 Front+Middle DNA | 12μL |
| 5 | CrdS No.15 Front+Middle DNA | 12μL |
| 6 | λ-HindⅢ | 10μL |
| 7 | 500bp DNA ladder | 10μL |
| 8 | - | - |
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS N Front+Rear DNA | 12μL |
| 4 | CrdS No.6 Front+Rear DNA | 12μL |
| 5 | CrdS No.15 Front+Rear DNA | 12μL |
| 6 | λ-HindⅢ | 10μL |
| 7 | 500bp DNA ladder | 10μL |
| 8 | - | - |
N, No.6, No.15 each fragment Overlap PCR completed.
Gel Purification of CrdS
No.8 DNA that about 2ooo bp band performed Gel Purification by illustra GFX PCR Purification Kit.
Adapter PCR of CrdS gene (BioBrick Part)
| No | Name | volume |
| 1 | 2-log DNA ladder | 5μL |
| 2 | λ-HindⅢ | 6μL |
| 3 | PCR saample | 18μL |
| 4 | - | - |
| 5 | - | - |
| 6 | - | - |
| 7 | - | - |
| 8 | - | - |
Fw Primer: GTTTCTTCGAATTCGCGGCCGCTTCTAGATG
Rv Primer: GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTATC
Brick Part of CrdS Restrict Enzyme,EcoRI and PstI.
| No | Name | volume |
| 1 | 500bp DNA ladder | 10μL |
| 2 | λ-HindⅢ | 10μL |
| 3 | CrdS N F+M non-RE | 12μL |
| 4 | CrdS N F+M EcoRⅠ-RE | 12μL |
| 5 | CrdS No.6 F+M non-RE | 12μL |
| 6 | CrdS No.6 F+M EcoRⅠ-RE | 12μL |
| 7 | CrdS No.15 F+M non-RE | 12μL |
| 8 | CrdS No.15 F+M EcoRⅠ | 12μL |
| 9 | CrdS N F+M+R non-RE | 12μL |
| 10 | CrdS N F+M+R PstⅠ-RE | 12μL |
| 11 | CrdS No.6 F+M+R non-RE | 12μL |
| 12 | Crds No.6 F+M+R PstI RE | 12μL |
| 13 | CrdS No.15 F+M+R non-RE | 12μL |
| 14 | CrdS No.15 F+M+R PstI RE | 12μL |
| 15 | λ-HindⅢ | 10μL |
| 16 | 500bp DNA ladder | 10μL |
| 17 | - | - |
| 18 | - | - |
 "
"