Team:Buenos Aires/ res basu
From 2013.igem.org
(→Experiment 1: hypoxia induced by anaerobiosis boxes) |
|||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
= Response of the construction [[http://parts.igem.org/Part:BBa_K1166000 Bba_K1166000]] under different hypoxia conditions = | = Response of the construction [[http://parts.igem.org/Part:BBa_K1166000 Bba_K1166000]] under different hypoxia conditions = | ||
| + | |||
| + | '''Objective''' | ||
We aimed to characterize the part sent by de Monterrey Team which is a GFP under a hypoxia inducible promoter, cloned in a replicative plasmid (Bba_K1166000). For that purpose we transformed E. coli DH5α with plamid Bba_K1166000 and conducted 2 different assays described below using this strain. | We aimed to characterize the part sent by de Monterrey Team which is a GFP under a hypoxia inducible promoter, cloned in a replicative plasmid (Bba_K1166000). For that purpose we transformed E. coli DH5α with plamid Bba_K1166000 and conducted 2 different assays described below using this strain. | ||
| Line 8: | Line 8: | ||
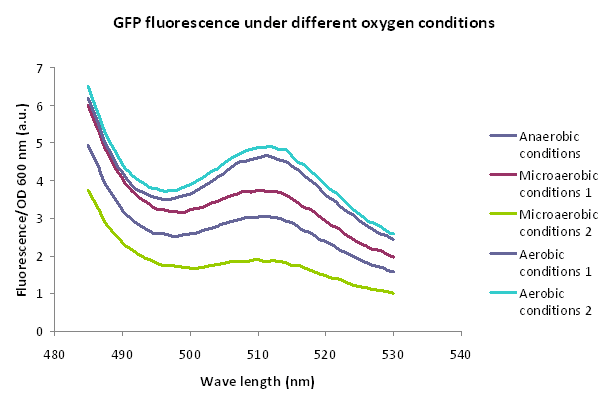
== '''Experiment 1: hypoxia induced by anaerobiosis boxes''' == | == '''Experiment 1: hypoxia induced by anaerobiosis boxes''' == | ||
| - | ''' | + | '''Method''' |
A 5ml culture of E. coli DH5α carrying the plasmid Bba_K1166000 in LB medium was grown for 16 h in aerobiosis, 1 ml was centrifuged and the pellet was kept frozen in order to use it as fluorescence control without induction. The rest of the culture was incubated for 20 h under anaerobic and microanaerobic conditions using hermetic boxes and GENbox aner and GENbox microanaer systems (bioMérieux), respectively, without shacking at 37°C. GENbox aner and GENbox microanaer systems are based on a chemical compound that traps oxygen. | A 5ml culture of E. coli DH5α carrying the plasmid Bba_K1166000 in LB medium was grown for 16 h in aerobiosis, 1 ml was centrifuged and the pellet was kept frozen in order to use it as fluorescence control without induction. The rest of the culture was incubated for 20 h under anaerobic and microanaerobic conditions using hermetic boxes and GENbox aner and GENbox microanaer systems (bioMérieux), respectively, without shacking at 37°C. GENbox aner and GENbox microanaer systems are based on a chemical compound that traps oxygen. | ||
| Line 14: | Line 14: | ||
| - | ''' | + | '''Results''' |
[[File:Hernanmexpng.PNG|center|600px]] | [[File:Hernanmexpng.PNG|center|600px]] | ||
| Line 20: | Line 20: | ||
== '''Experiment 2: hipoxia induced in a microscope slide sealed''' == | == '''Experiment 2: hipoxia induced in a microscope slide sealed''' == | ||
| - | ''' | + | '''Method''' |
An E. coli DH5α carrying the plasmid Bba_K1166000 was grown under aerobic conditions overnight. Then, 10ul of this culture was placed in a microscope slide, covered with a coverslip and sealed with nail enamel (similar as usually performed to induce hypoxia in plant cells). | An E. coli DH5α carrying the plasmid Bba_K1166000 was grown under aerobic conditions overnight. Then, 10ul of this culture was placed in a microscope slide, covered with a coverslip and sealed with nail enamel (similar as usually performed to induce hypoxia in plant cells). | ||
| Line 26: | Line 26: | ||
Afterwards, pictures of bacteria were taken in a fluorescence microscope every 15 min for 1 h. The first picture was taken immediately after sealing. Every picture was taken using the same exposition time (1/5 sec). Unfortunately, a lot of bleaching was observed. To avoid this, the field was changed before taking each photo. From 0 min, bacteria exhibited green fluorescence, however no significant induction can be observed in the time points analyzed (15, 30, 45 and 60 min). | Afterwards, pictures of bacteria were taken in a fluorescence microscope every 15 min for 1 h. The first picture was taken immediately after sealing. Every picture was taken using the same exposition time (1/5 sec). Unfortunately, a lot of bleaching was observed. To avoid this, the field was changed before taking each photo. From 0 min, bacteria exhibited green fluorescence, however no significant induction can be observed in the time points analyzed (15, 30, 45 and 60 min). | ||
| - | ''' | + | '''Results''' |
[[File:Carlotto1.JPG|center|600px]] | [[File:Carlotto1.JPG|center|600px]] | ||
Revision as of 02:23, 28 September 2013
Response of the construction [Bba_K1166000] under different hypoxia conditions
Objective
We aimed to characterize the part sent by de Monterrey Team which is a GFP under a hypoxia inducible promoter, cloned in a replicative plasmid (Bba_K1166000). For that purpose we transformed E. coli DH5α with plamid Bba_K1166000 and conducted 2 different assays described below using this strain.
Experiment 1: hypoxia induced by anaerobiosis boxes
Method
A 5ml culture of E. coli DH5α carrying the plasmid Bba_K1166000 in LB medium was grown for 16 h in aerobiosis, 1 ml was centrifuged and the pellet was kept frozen in order to use it as fluorescence control without induction. The rest of the culture was incubated for 20 h under anaerobic and microanaerobic conditions using hermetic boxes and GENbox aner and GENbox microanaer systems (bioMérieux), respectively, without shacking at 37°C. GENbox aner and GENbox microanaer systems are based on a chemical compound that traps oxygen. One ml of each condition was centrifuged and the pellets frozen. Finally, fluorescence (excitation: 475 nm) was measure and normalized by OD600nm. Both treatments were performed in duplicate.
Results
Experiment 2: hipoxia induced in a microscope slide sealed
Method
An E. coli DH5α carrying the plasmid Bba_K1166000 was grown under aerobic conditions overnight. Then, 10ul of this culture was placed in a microscope slide, covered with a coverslip and sealed with nail enamel (similar as usually performed to induce hypoxia in plant cells).
Afterwards, pictures of bacteria were taken in a fluorescence microscope every 15 min for 1 h. The first picture was taken immediately after sealing. Every picture was taken using the same exposition time (1/5 sec). Unfortunately, a lot of bleaching was observed. To avoid this, the field was changed before taking each photo. From 0 min, bacteria exhibited green fluorescence, however no significant induction can be observed in the time points analyzed (15, 30, 45 and 60 min).
Results
</div>
 "
"