Team:DTU-Denmark/HelloWorld
From 2013.igem.org
(→Methods) |
(→Methods) |
||
| Line 10: | Line 10: | ||
== Methods == | == Methods == | ||
| - | [[File:Both.png| | + | [[File:Both.png|680px]] |
Revision as of 22:44, 1 October 2013
Hello World Pilot Project
Contents |
Introduction
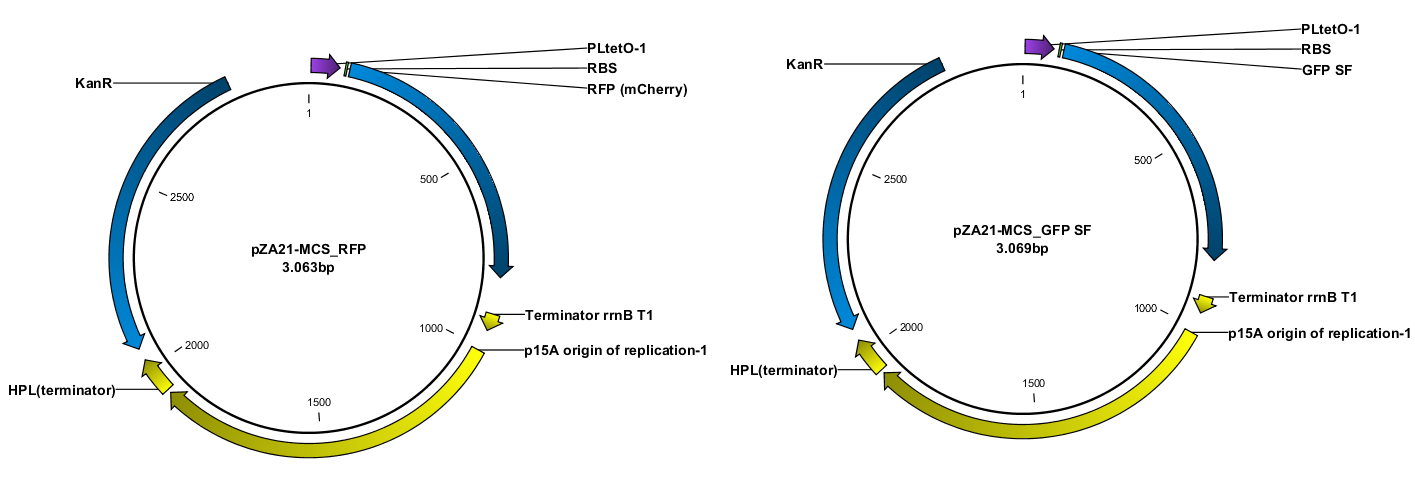
‘Hello World!’ are the first words a programmer prints when learning a new programming language. In analogy to this our team decided to do a ‘Hello World’ project in order to familiarize ourselves with lab techniques that we used later on to construct plasmids. Specifically we were performing PCR with uracil-containing primers, purifying PCR products and ligating them by means of USER cloning (Nour-Eldin, H. H.).
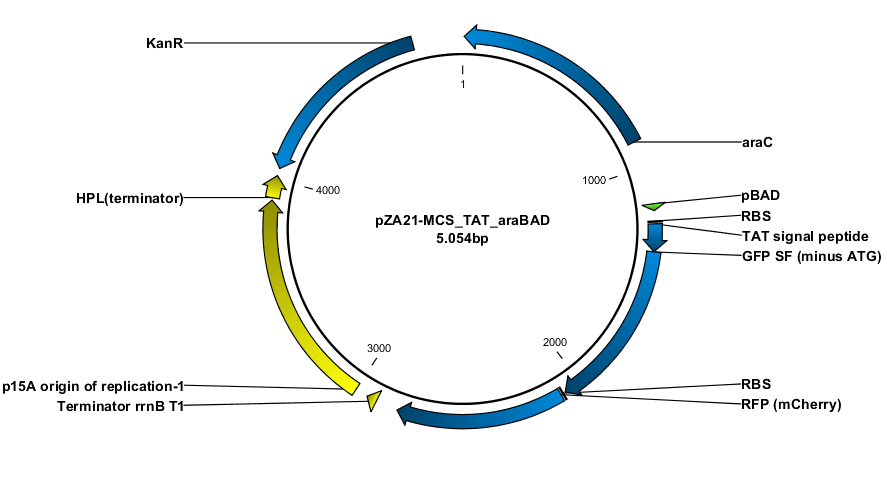
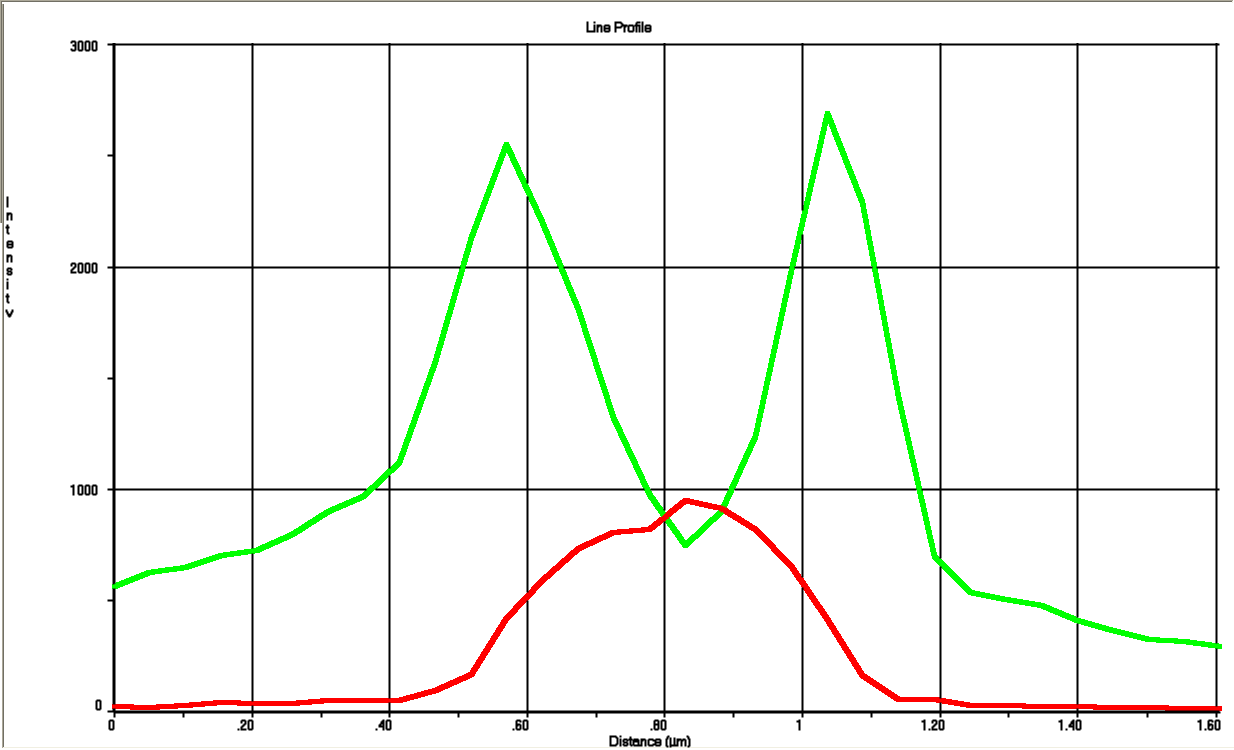
Since we are working with many periplasmic proteins, we wanted to try to target proteins to the periplasm. To do this, we used periplasmic signal peptides from the TAT and Sec pathways, and with a translational fusion of the signal peptide to GFP, we expressed GFP in the periplasm. Simultaneously, we expressed RFP in the cytoplasm as a background color, inspired by Skoog, Karl, et al.
Methods
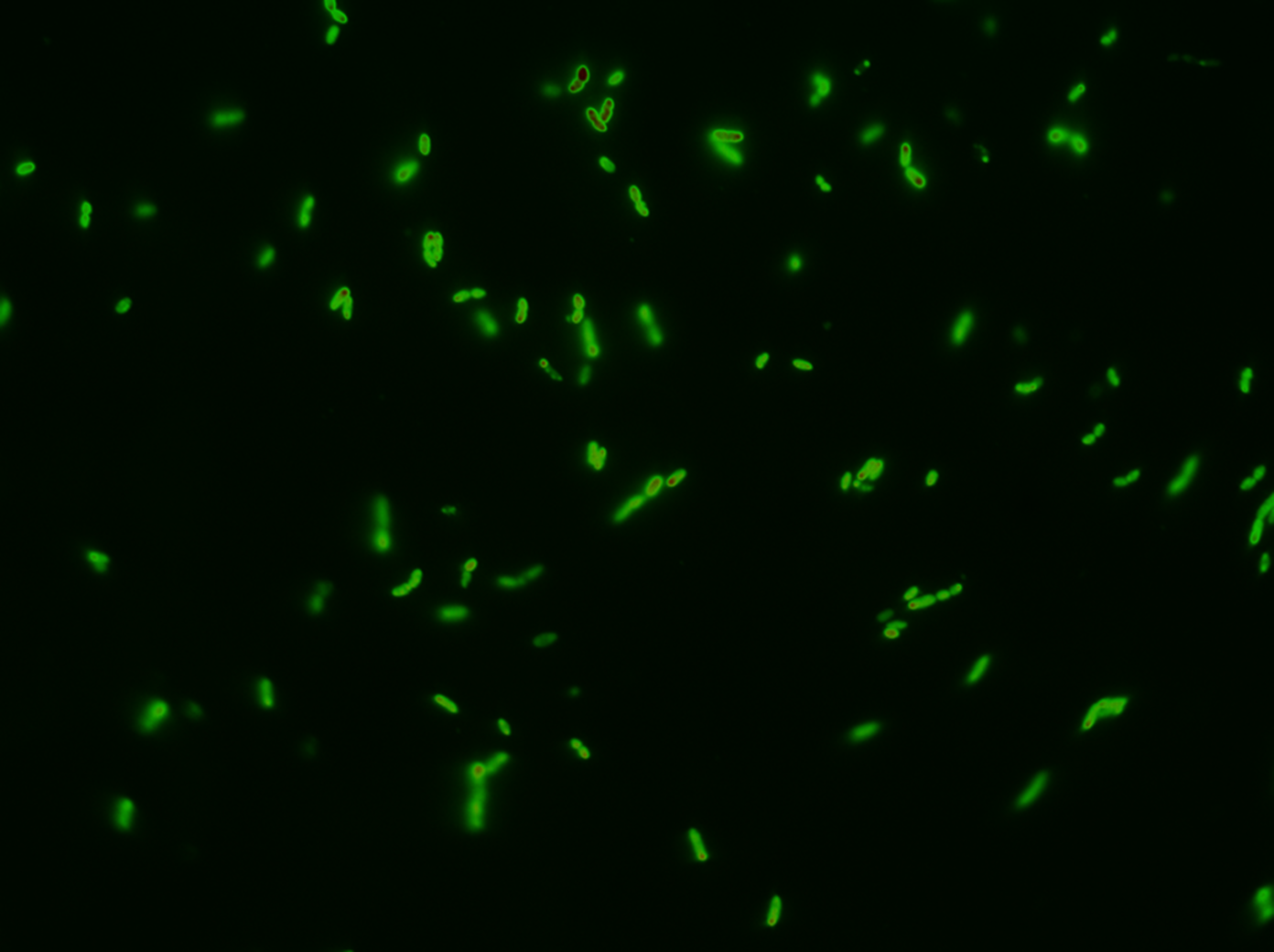
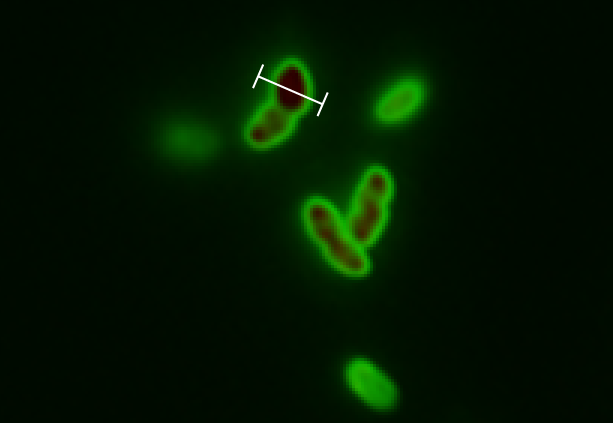
We followed this protocol to visualize GFP in the periplasm.
Results
Conclusions
Biobrick BBa_K1067009 successfully directs proteins to the periplasm in E. coli.
References
- Skoog, Karl, et al. "Sequential Closure of the Cytoplasm and Then the Periplasm during Cell Division in Escherichia coli." Journal of bacteriology 194.3 (2012): 584-586.
- Nour-Eldin, H. H., Geu-Flores, F., & Halkier, B. A. (2010). USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. In Plant Secondary Metabolism Engineering (pp. 185-200). Humana Press.
 "
"