Team:DTU-Denmark/Notebook/12 July 2013
From 2013.igem.org
(→208) |
|||
| Line 1: | Line 1: | ||
| + | {{:Team:DTU-Denmark/Templates/StartPage|13 July 2013}} | ||
| + | __TOC__ | ||
| + | |||
| + | |||
=208= | =208= | ||
| + | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
| + | <hr/> | ||
*miniprep of biobrick transformants from [[Team:DTU-Denmark/Notebook/10_July_2013#Transformation_of_Biobricks|10-07-2013]] | *miniprep of biobrick transformants from [[Team:DTU-Denmark/Notebook/10_July_2013#Transformation_of_Biobricks|10-07-2013]] | ||
*PCR with USER primers to amplify cytochromes, AMO, HAO and Nir genes | *PCR with USER primers to amplify cytochromes, AMO, HAO and Nir genes | ||
*gel analysis of PCR products from [[Team:DTU-Denmark/Notebook/11_July_2013#Combine_Nir_operon_with_pZA21|Nir operon]], [[Team:DTU-Denmark/Notebook/11_July_2013#PCR_for_promoter_test| AraBAD promoter]] and [[Team:DTU-Denmark/Notebook/11_July_2013#PCR_for_promoter_test| RFP]] | *gel analysis of PCR products from [[Team:DTU-Denmark/Notebook/11_July_2013#Combine_Nir_operon_with_pZA21|Nir operon]], [[Team:DTU-Denmark/Notebook/11_July_2013#PCR_for_promoter_test| AraBAD promoter]] and [[Team:DTU-Denmark/Notebook/11_July_2013#PCR_for_promoter_test| RFP]] | ||
* USER reaction of [[Team:DTU-Denmark/Notebook/11_July_2013#PCR_for_promoter_test| RFP]] and pZA21 (with native promoter) and transformation of ''E. coli'' cells | * USER reaction of [[Team:DTU-Denmark/Notebook/11_July_2013#PCR_for_promoter_test| RFP]] and pZA21 (with native promoter) and transformation of ''E. coli'' cells | ||
| + | *Gel on todays PCR-reactions. | ||
==Who was in the lab== | ==Who was in the lab== | ||
| + | <hr/> | ||
Henrike, Julia, Kristian, Jakob, Gosia | Henrike, Julia, Kristian, Jakob, Gosia | ||
==Procedure== | ==Procedure== | ||
| + | <hr/> | ||
===Plasmid isolation of biobricks from transformants made on [[Team:DTU-Denmark/Notebook/10_July_2013|10-07-2013]]=== | ===Plasmid isolation of biobricks from transformants made on [[Team:DTU-Denmark/Notebook/10_July_2013|10-07-2013]]=== | ||
| + | <hr/> | ||
Isolation was performed according to protocol given in the kit PowerPrep HP Plasmid Miniprep Kit | Isolation was performed according to protocol given in the kit PowerPrep HP Plasmid Miniprep Kit | ||
===Amplification of cytochromes, AMO, HAO and Nir genes=== | ===Amplification of cytochromes, AMO, HAO and Nir genes=== | ||
| + | <hr/> | ||
Each PCR reaction was performed in triplicate. | Each PCR reaction was performed in triplicate. | ||
| Line 38: | Line 49: | ||
===Gel analysis of PCR products from Nir operon, AraBAD promoter and RFP === | ===Gel analysis of PCR products from Nir operon, AraBAD promoter and RFP === | ||
| - | + | <hr/> | |
1 % Agorase Gel (Nir operon) | 1 % Agorase Gel (Nir operon) | ||
Wells | Wells | ||
| - | *1: ladder 100bp | + | *1: ladder 100bp NEB |
*2: RFP | *2: RFP | ||
*3: RFP | *3: RFP | ||
| Line 62: | Line 73: | ||
===USER reaction of RFP and pZA21 (with native promoter) and transformation of E. coli cells=== | ===USER reaction of RFP and pZA21 (with native promoter) and transformation of E. coli cells=== | ||
| - | + | <hr/> | |
USER reaction was performed according to standard protocol. | USER reaction was performed according to standard protocol. | ||
| Line 81: | Line 92: | ||
Incubation of cells on ice for 30 minutes, heat shock at 42C for 90 sec. and 5 min on ice. Addition of SOC medium and 2 hours of incubation at 37C. Plating on plates with LB medium with Kanamycin, overnight incubation at 37C. | Incubation of cells on ice for 30 minutes, heat shock at 42C for 90 sec. and 5 min on ice. Addition of SOC medium and 2 hours of incubation at 37C. Plating on plates with LB medium with Kanamycin, overnight incubation at 37C. | ||
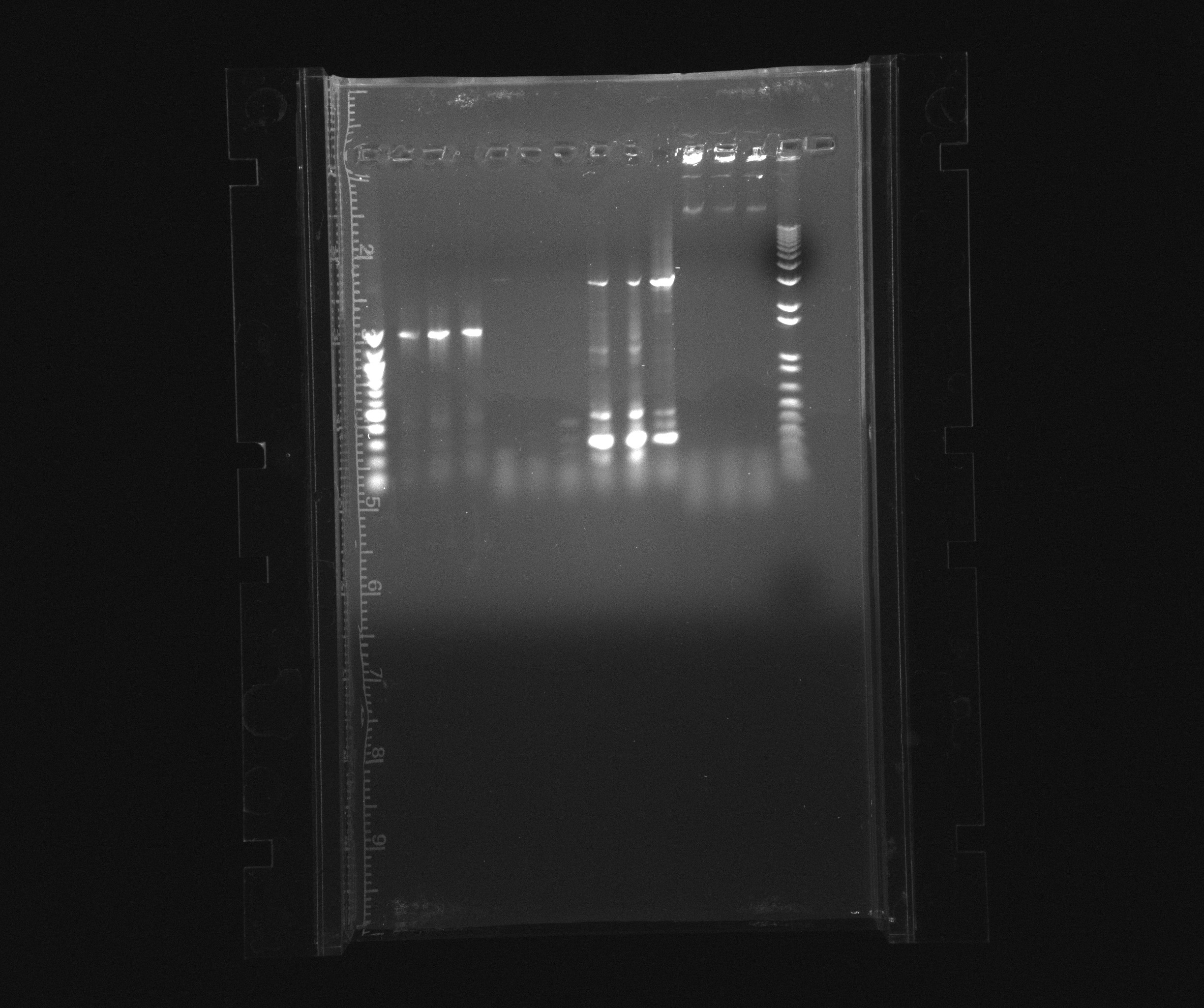
| + | ===Gel 2; on todays PCR-products for USER-cloning (cycAX, AMO, HAO, Nir)=== | ||
| + | <hr/> | ||
| + | Wells | ||
| + | *1: ladder 100bp NEB | ||
| + | *2: cycAX, tube 1 | ||
| + | *3: cycAX tube 2 | ||
| + | *4: cycAX tube 3 | ||
| + | *5: AMO tube 4 | ||
| + | *6: AMO tube 5 | ||
| + | *7: AMO tube 6 | ||
| + | *8: HAO tube 7 | ||
| + | *9: HAO tube 8 | ||
| + | *10: HAO tube 9 | ||
| + | *11: Nir tube 10 | ||
| + | *12: Nir tube 11 | ||
| + | *13: Nir tube 12 | ||
| + | *14: Ladder 1 kb | ||
| + | [[File:2013 07 12 gel USER cycAX AMO HAO nir.jpg| 600px]] | ||
| + | |||
| + | |||
| + | ====Colony PCR to extract the Nir operon directly with USER-primers==== | ||
| + | <hr/> | ||
==Conclusion== | ==Conclusion== | ||
| - | We have USER fragments for RFP and AraBAD but Nir operon is another story. | + | <hr/> |
| - | The Nir primers seems to make lots of unspecific products. We | + | We have USER fragments for RFP and AraBAD which was verified with the first gel but Nir operon is another story. |
| + | The Nir primers seems to make lots of unspecific products. We tried to do the colony PCR with the USER primers directly and this gave fragments longer than expected but not with unspecific product though. We will try and transform this fragment into E.coli and see what will happen and then we will send the construct to sequencing so that we can see whats causing the elongation of the fragment. | ||
| + | |||
| + | |||
| + | |||
| + | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Revision as of 13:26, 15 July 2013
13 July 2013
208
Main purpose
- miniprep of biobrick transformants from 10-07-2013
- PCR with USER primers to amplify cytochromes, AMO, HAO and Nir genes
- gel analysis of PCR products from Nir operon, AraBAD promoter and RFP
- USER reaction of RFP and pZA21 (with native promoter) and transformation of E. coli cells
- Gel on todays PCR-reactions.
Who was in the lab
Henrike, Julia, Kristian, Jakob, Gosia
Procedure
Plasmid isolation of biobricks from transformants made on 10-07-2013
Isolation was performed according to protocol given in the kit PowerPrep HP Plasmid Miniprep Kit
Amplification of cytochromes, AMO, HAO and Nir genes
Each PCR reaction was performed in triplicate.
Samples are named:
- 1, 2, 3 -> cytochromes
- 4, 5, 6 -> AMO
- 7, 8, 9 -> HAO
- 10, 11, 12 -> Nir
PCR reaction mix was done according to standard procedure.Primers and templates used for samples are as follows:
- 1, 2, 3 -> cytochromes, primers 16a, 16b, template CYC isolated by colony PCR from Nitrosomonas europea
- 4, 5, 6 -> AMO, primers 17a, 17b, template AMO isolated by colony PCR from N. europea
- 7, 8, 9 -> HAO, primers 18a, 18b, template HAO isolated by colony PCR from N. europea
- 10, 11, 12 -> Nir, primers 15a, 15b, template - 2uL of liquid culture of "Pseudosomonas"
PCR programs based on standard program with differences in annealing temperature and elongation time as follows:
- 1, 2, 3 -> cytochromes - 57°C, 1:30 min
- 4, 5, 6 -> AMO - 54°C, 3 min
- 7, 8, 9 -> HAO - 59°C, 9 min
- 10, 11, 12 -> Nir - 59°C, 9 min
Gel analysis of PCR products from Nir operon, AraBAD promoter and RFP
1 % Agorase Gel (Nir operon)
Wells
- 1: ladder 100bp NEB
- 2: RFP
- 3: RFP
- 4: AraBAD promoter
- 5: AraBAD promoter
- 6: AraBAD promoter
- 7: RFP
- 8: Nir
- 9: Nir
- 10: Nir
- 11:Nir
- 12: Nir
- 13: Nir
- 14: Nir
- 15: Ladder 1 kb
USER reaction of RFP and pZA21 (with native promoter) and transformation of E. coli cells
USER reaction was performed according to standard protocol.
USER mix:
- USER enzyme 1 uL
- NEB 4 buffer 0,5 uL
- 10x BSA 0,5 uL
- backbone pZA21 1uL
- Sample 1 - 3 uL of USER mix + 7 uL of RFP
- Sample 2 - Negative control, USER mix + 7 uL water
USER reaction was performed in PCR machine with programm: 37C for 40 min and then 25C for 30 min.
To 100uL of chemically competent E. coli cells 10 uL of USER mix after USER reaction was added.
Incubation of cells on ice for 30 minutes, heat shock at 42C for 90 sec. and 5 min on ice. Addition of SOC medium and 2 hours of incubation at 37C. Plating on plates with LB medium with Kanamycin, overnight incubation at 37C.
Gel 2; on todays PCR-products for USER-cloning (cycAX, AMO, HAO, Nir)
Wells
- 1: ladder 100bp NEB
- 2: cycAX, tube 1
- 3: cycAX tube 2
- 4: cycAX tube 3
- 5: AMO tube 4
- 6: AMO tube 5
- 7: AMO tube 6
- 8: HAO tube 7
- 9: HAO tube 8
- 10: HAO tube 9
- 11: Nir tube 10
- 12: Nir tube 11
- 13: Nir tube 12
- 14: Ladder 1 kb
Colony PCR to extract the Nir operon directly with USER-primers
Conclusion
We have USER fragments for RFP and AraBAD which was verified with the first gel but Nir operon is another story. The Nir primers seems to make lots of unspecific products. We tried to do the colony PCR with the USER primers directly and this gave fragments longer than expected but not with unspecific product though. We will try and transform this fragment into E.coli and see what will happen and then we will send the construct to sequencing so that we can see whats causing the elongation of the fragment.
 "
"