Team:DTU-Denmark/Notebook/31 July 2013
From 2013.igem.org
(→lab ...) |
|||
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|31 July 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|31 July 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/30_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/1_August_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| + | |||
| + | * USER reaction to integrate Biobrick K808000 (AraBAD promoter system) in pZE21 containing fluorescence markers | ||
| + | * PCR | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 12: | Line 15: | ||
==Procedure== | ==Procedure== | ||
<hr/> | <hr/> | ||
| + | |||
| + | ===USER=== | ||
| + | |||
| + | USER reaction to integrate Biobrick K808000 (AraBAD promoter system) in pZE21 containing either GFP SF or RFP. Did doubles for both with 7 uL of AraBAD and 15 uL to compare. Standard procedure. | ||
| + | |||
| + | ===PCR=== | ||
| + | Performed PCR on lambda phage as a test of polymerase efficiency. (PHUSION polymerase test kit) | ||
| + | |||
| + | PCR on HAO and AMO with USER primer pair 17 with 3:00 on 54C and 18 with 3:00 on 59C respectively. Tried out both colonies (col) and PCR DNA template (pur): | ||
| + | *HAO pur | ||
| + | *HAO pur | ||
| + | *HAO col | ||
| + | *HAO col | ||
| + | *AMO pur | ||
| + | *AMO pur | ||
| + | *AMO col | ||
| + | *AMO col | ||
| + | |||
| + | |||
| + | Later on the day we made another PCR on cycAX with his-tag 5 rxn with primer pair 33 and 3:30 min extension time. This time with lower temperature: | ||
| + | *2x on 50C | ||
| + | *2x on 45C | ||
| + | *1x neg. on 45C | ||
==Results== | ==Results== | ||
| Line 21: | Line 47: | ||
* HAO col | * HAO col | ||
* HAO col | * HAO col | ||
| - | * HAO | + | * HAO pur |
| - | * HAO | + | * HAO pur |
* AMO col | * AMO col | ||
* AMO col | * AMO col | ||
| - | * AMO | + | * AMO pur |
| - | * AMO | + | * AMO pur |
* neg | * neg | ||
* 1 kb ladder | * 1 kb ladder | ||
| - | * | + | * polymerase test with PHUSION |
| + | * polymerase test with PHUSION | ||
| + | * polymerase test with X7 | ||
| + | * polymerase test with X7 | ||
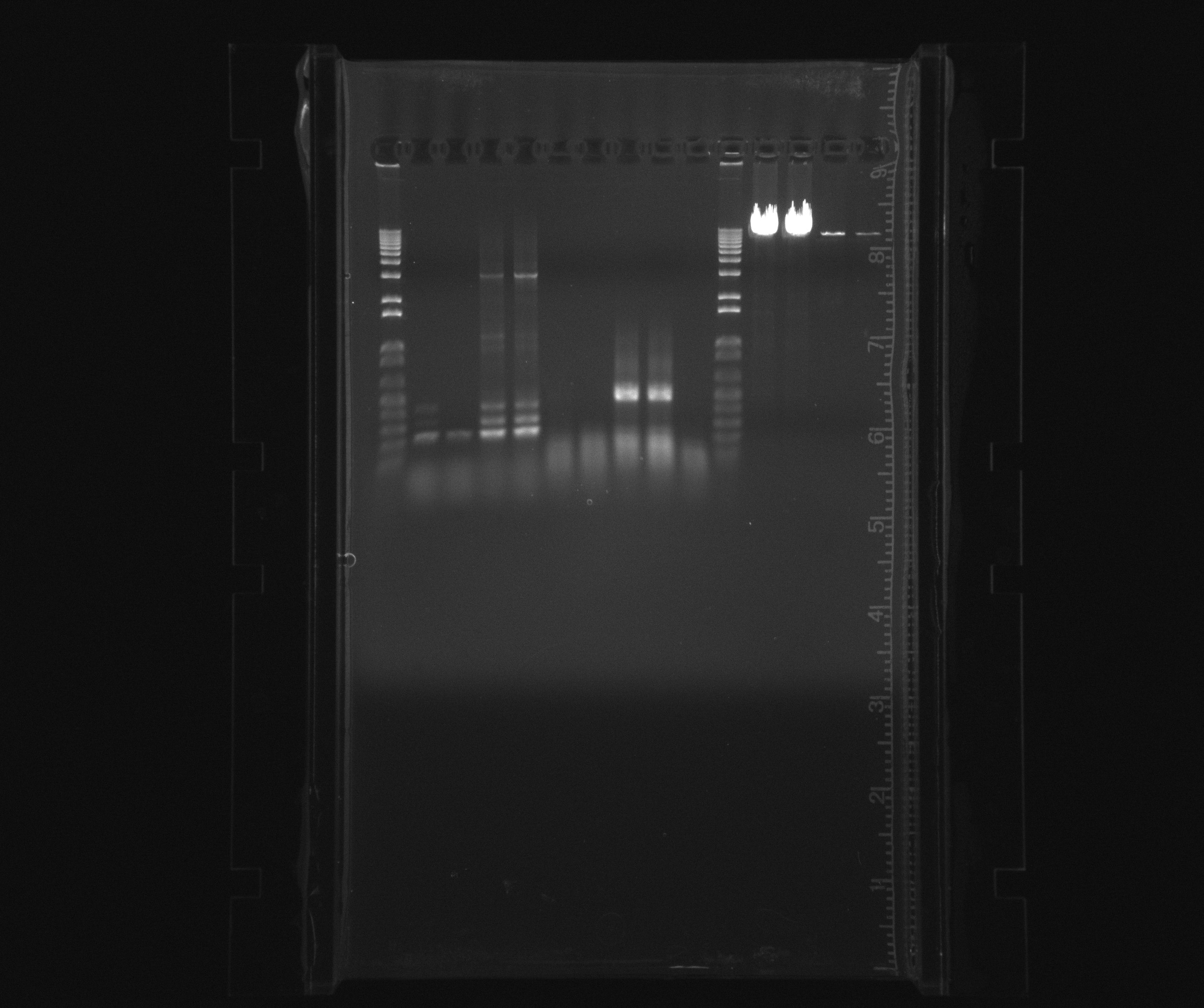
| + | [[File:2013-07-31 hao amo test.jpg|600px]] | ||
==Conclusion== | ==Conclusion== | ||
<hr/> | <hr/> | ||
| + | The positive control was good with both cases of polymerase. | ||
| + | |||
Navigate to the [[Team:DTU-Denmark/Notebook/30_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/1_August_2013|Next]] Entry | Navigate to the [[Team:DTU-Denmark/Notebook/30_July_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/1_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 20:45, 16 September 2013
31 July 2013
Contents |
Lab 208
Main purpose
- USER reaction to integrate Biobrick K808000 (AraBAD promoter system) in pZE21 containing fluorescence markers
- PCR
Who was in the lab
Kristian, Gosia, Julia, Henrike
Procedure
USER
USER reaction to integrate Biobrick K808000 (AraBAD promoter system) in pZE21 containing either GFP SF or RFP. Did doubles for both with 7 uL of AraBAD and 15 uL to compare. Standard procedure.
PCR
Performed PCR on lambda phage as a test of polymerase efficiency. (PHUSION polymerase test kit)
PCR on HAO and AMO with USER primer pair 17 with 3:00 on 54C and 18 with 3:00 on 59C respectively. Tried out both colonies (col) and PCR DNA template (pur):
- HAO pur
- HAO pur
- HAO col
- HAO col
- AMO pur
- AMO pur
- AMO col
- AMO col
Later on the day we made another PCR on cycAX with his-tag 5 rxn with primer pair 33 and 3:30 min extension time. This time with lower temperature:
- 2x on 50C
- 2x on 45C
- 1x neg. on 45C
Results
Gel on todays PCR
- 1kb ladder
- HAO col
- HAO col
- HAO pur
- HAO pur
- AMO col
- AMO col
- AMO pur
- AMO pur
- neg
- 1 kb ladder
- polymerase test with PHUSION
- polymerase test with PHUSION
- polymerase test with X7
- polymerase test with X7
Conclusion
The positive control was good with both cases of polymerase.
Navigate to the Previous or the Next Entry
 "
"