Team:DTU-Denmark/Project
From 2013.igem.org
(→Mutant 2: Anaerobic) |
|||
| (69 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
__NOTOC__ | __NOTOC__ | ||
==Project Description== | ==Project Description== | ||
| + | ===What?=== | ||
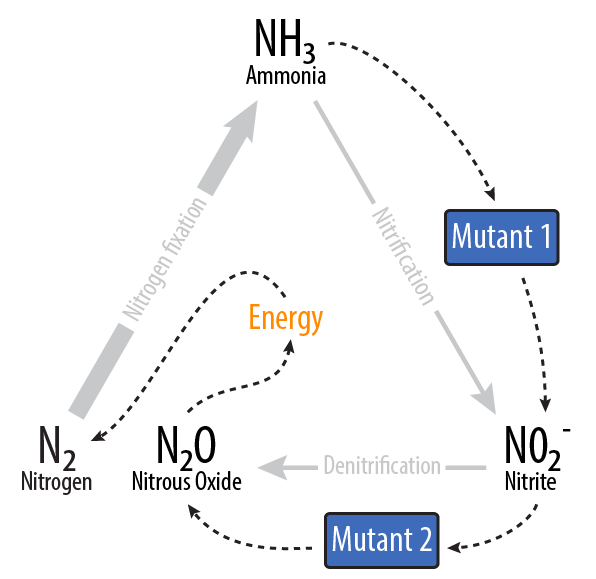
| + | Our project removes ammonia (NH<sub>3</sub>) from waste water, and via two ''E. coli'' mutants, turns it into nitrous oxide (N<sub>2</sub>O). | ||
| - | + | [[File:Dtu overview.png|400px|center]] | |
| - | Global demand for fixed nitrogen has increased to the point that half the human population now relies on chemical fertilizer to grow their food. While fertilizer is a requirement for modern life, runoff from overfertilized farmland can cause eutrophication. In the presence of abundant ammonia, algae overgrow and consume the much of the available oxygen in the water. This results in decreased biodiversity throughout the watershed. Within Europe, 53% of lakes are eutrophic. | + | ===Why?=== |
| + | Global demand for fixed nitrogen has increased to the point that half the human population now relies on chemical fertilizer to grow their food. While fertilizer is a requirement for modern life, runoff from overfertilized farmland can cause eutrophication. In the presence of abundant ammonia, algae overgrow and consume the much of the available oxygen in the water [1]. This results in decreased biodiversity throughout the watershed. Within Europe, 53% of lakes are eutrophic. | ||
| - | Using two ''E. coli'' mutants built with genes from ''Nitrosomonas europaea'' and ''Pseudomonas aeruginosa'', we provide a system to reverse nitrogen fixation. Our mutants consume ammonia and produce nitrous oxide, releases a sustainable source of energy when decomposed into nitrogen and oxygen. We also provide a | + | In Europe the annual surplus of fixed nitrogen from crop production and livestock farming is 10 Tg (10 Teragrams = a million tonnes), 6 Tg of which leaches into ground and surface water [2]. If that was to be converted into N<sub>2</sub>O, it could be decomposed for 4.87 Billion kW h -- equivalent to the annual power consumption of 36,000 Avarage households. |
| + | |||
| + | If released into the atmosphere, nitrous oxide is a potent greenhouse gas, and capable of degrading ozone, which is why it makes sense to convert it to N<sub>2</sub>. | ||
| + | |||
| + | ===How?=== | ||
| + | Using two ''E. coli'' mutants built with genes from ''Nitrosomonas europaea'' and ''Pseudomonas aeruginosa'', we provide a system to reverse nitrogen fixation. Our mutants consume ammonia and produce nitrous oxide, releases a sustainable source of energy when decomposed into nitrogen and oxygen [3]. We also provide [[Team:DTU-Denmark/Reactor_Model|a model of a bioreactor]] that could be scaled up and deployed in the field to simultaneously clean the water and produce energy. | ||
==Details== | ==Details== | ||
=== Mutant 1: Aerobic === | === Mutant 1: Aerobic === | ||
| + | |||
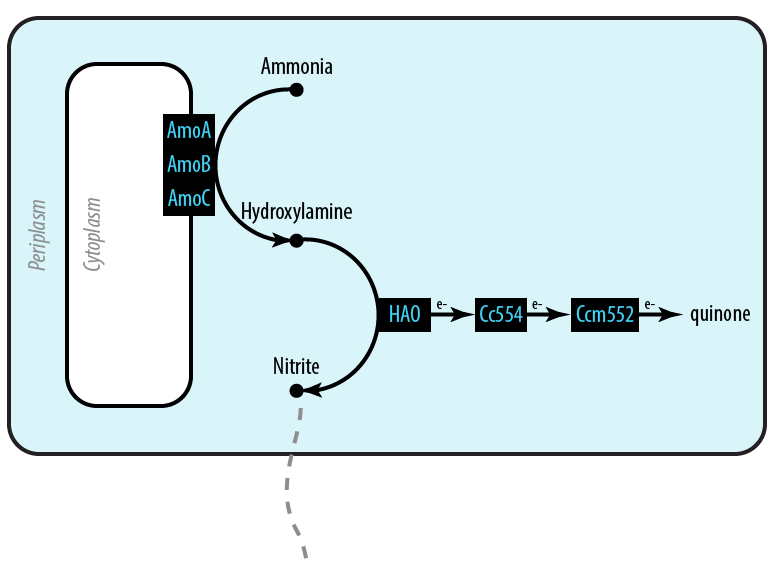
[[File:Dtu-pathway-mutant1.png]] | [[File:Dtu-pathway-mutant1.png]] | ||
| - | The first mutant incorporates [[Team:DTU-Denmark/Protein_Models#AMO|AMO]], [[Team:DTU-Denmark/Protein_Models#Hao|HAO]] and two cytochromes [[Team:DTU-Denmark/Protein_Models#Cc554|c554]] and [[Team:DTU-Denmark/Protein_Models#Ccm552|cm552]] from ''Nitrosomonas europaea'' (shown in blue). AMO is a 3 subunit protein which converts ammonia (NH<sub>4</sub> to an intermediate called hydroxylamine (NH<sub>2</sub>OH). HAO then converts the hydroxylamine to nitrite (NO<sub>2</sub><sup>-</sup>. During this conversion process, cytochrome c554 accepts an electron from HAO, and then passes this electron on to ccm552. The terminal electron acceptor in this chain is quinone. AMO and ccm552 are embedded in the cytoplasmic membrane, and all other proteins in this process are found in the periplasm. | + | [[File:Dtu-Mutant1.png|right]]The first mutant incorporates [[Team:DTU-Denmark/Protein_Models#AMO|AMO]], [[Team:DTU-Denmark/Protein_Models#Hao|HAO]] and two cytochromes [[Team:DTU-Denmark/Protein_Models#Cc554|c554]] and [[Team:DTU-Denmark/Protein_Models#Ccm552|cm552]] from ''Nitrosomonas europaea'' (shown in blue). AMO is a 3 subunit protein which converts ammonia (NH<sub>4</sub>) to an intermediate called hydroxylamine (NH<sub>2</sub>OH). HAO then converts the hydroxylamine to nitrite (NO<sub>2</sub><sup>-</sup>). During this conversion process, cytochrome c554 accepts an electron from HAO, and then passes this electron on to ccm552. The terminal electron acceptor in this chain is quinone. AMO and ccm552 are embedded in the cytoplasmic membrane, and all other proteins in this process are found in the periplasm. |
| + | <br> | ||
| + | <br> | ||
=== Mutant 2: Anaerobic === | === Mutant 2: Anaerobic === | ||
| + | |||
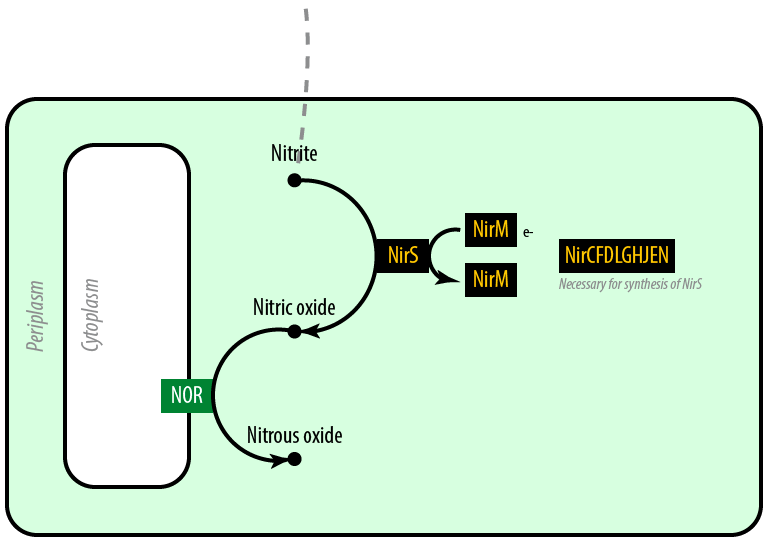
[[File:Dtu-pathway-mutant2.png]] | [[File:Dtu-pathway-mutant2.png]] | ||
| - | The second mutant incorporates the Nir region from ''Pseudomonas aeruginosa'' (shown in yellow). [[Team:DTU-Denmark/Protein_Models#NirS|NirS]] converts nitrite to nitric oxide (NO), while removing an electron from [[Team:DTU-Denmark/Protein_Models#NirM|NirM]]. The remainder of the Nir region is necessary for the synthesis of NirS, and so we have included these genes as well. [[Team:DTU-Denmark/Protein_Models#NOR|NOR]], which is present natively in ''E. coli'', converts nitric oxide to nitrous oxide (N<sub>2</sub>O). In contrast to ''N. europaea'', ''E. coli'' is not a nitrifying bacterium and so does not convert nitrous oxide into nitrogen to complete the denitrification process. | + | [[File:dtu-Mutant2.png|right]]The second mutant incorporates the Nir region from ''Pseudomonas aeruginosa'' (shown in yellow). [[Team:DTU-Denmark/Protein_Models#NirS|NirS]] converts nitrite to nitric oxide (NO), while removing an electron from [[Team:DTU-Denmark/Protein_Models#NirM|NirM]]. The remainder of the Nir region is necessary for the synthesis of NirS, and so we have included these genes as well. [[Team:DTU-Denmark/Protein_Models#NOR|NOR]], which is present natively in ''E. coli'', converts nitric oxide to nitrous oxide (N<sub>2</sub>O). In contrast to ''N. europaea'', ''E. coli'' is not a nitrifying bacterium and so does not convert nitrous oxide into nitrogen to complete the denitrification process. |
== Plasmids == | == Plasmids == | ||
| Line 26: | Line 38: | ||
=== Mutant 1 === | === Mutant 1 === | ||
| - | The final plasmid for Mutant 1 should be the following: | + | The final plasmid for Mutant 1 should be the following which includes all genes on one plasmid: |
| - | [[File:dtu_Plasmid_mutant_1-01.png| | + | [[File:dtu_Plasmid_mutant_1-01.png|300px]] |
| - | In order to simplify and to be able to debug each part of this pathway separately, we implemented the genes individually. | + | In order to simplify and to be able to debug each part of this pathway separately, we first implemented the genes individually. Unfortunately, we did not have time to combine all the genes into the final plasmid, but we were able to [[Team:DTU-Denmark/Experiment4|verify that the AMO containing plasmid is working as designed]]. |
| - | + | [[File:dtu-mutant1-implementation.png|600px]] | |
| - | + | ||
| - | [[File: | + | |
=== Mutant 2 === | === Mutant 2 === | ||
| - | The final plasmid for Mutant 2 | + | The final plasmid for Mutant 2 is the following: |
| - | [[File:Dtu Plasmid mutant 2-01.png| | + | [[File:Dtu Plasmid mutant 2-01.png|300px]] |
| - | Since we had difficulty extracting Nir from ''Pseudomonas aeruginosa'' in one piece, we split it into two pieces to be extracted seperately and then | + | Since we had difficulty extracting Nir from ''Pseudomonas aeruginosa'' in one piece, we split it into two pieces to be extracted seperately and then re-combined them with USER cloning. |
| - | |||
| - | To assemble our plasmids we employ uracil-specific excision reagent (USER) cloning | + | == USER Cloning == |
| + | To assemble our plasmids we employ uracil-specific excision reagent (USER) cloning [4]. This technique enables us to speed up the cloning process, to clone seamlessly, and to assemble many fragments in one reaction. The steps are: | ||
| - | # PCR: The backbone and all fragments are amplified as linear products containing a uracil instead of a thymine at a specific position. This requires | + | # PCR: The backbone and all fragments are amplified as linear products containing a uracil instead of a thymine at a specific position. This requires specially designed set of primers with a tail that will become the overhang and a polymerase that is able to perform uracil-insertion. |
| - | # Optional - If | + | ## We are using the non-commercial polymerase X7, designed by Morten Nørholm [5]. |
| - | # USER-reaction: Digesting the linearized PCR fragment with | + | # Optional - If the method was used to insert genes into a backbone, with it's own resistance marker, the PCR product were treated with DpnI to digest the template DNA from the PCR reaction. |
| - | # Transformation: The mix is immediately transformed into competent E. coli cells | + | # USER-reaction: Digesting the linearized PCR fragment with Uracil-Specific Excision Reagent (USER) enzyme removes the uracil-insertions. This will produce the required overhangs with the sequence and the length of your wish. |
| + | # Transformation: The mix is immediately transformed into competent ''E. coli'' cells. The plasmid assembles itself by base pairing between the complementary overhangs and ligation is done by ''E.coli'' itself. | ||
| - | See our "Bricks of Knowledge" [https://www.youtube.com/watch?v=7EiVttJpXH4 video on USER cloning] and the [https://2011.igem.org/Team:DTU-Denmark-2 wiki of the 2011 DTU-2 iGEM team] for more information. | + | '''Advantages to USER cloning''' |
| + | *You only need one enzyme | ||
| + | *You don't need any ligation reaction | ||
| + | *Seamless assembly | ||
| + | *Directional | ||
| + | *Multiple fragment in one reaction - also in eukaryotes [6]. | ||
| + | *[https://2011.igem.org/Team:DTU-Denmark-2 Plug 'n' Play] | ||
| + | |||
| + | [[File:Plug 'n' Play assembly samlet.png|thumb|upright=3|center|The [https://2011.igem.org/Team:DTU-Denmark-2 Plug 'n' Play] concept. Figure taken from DTU-2 iGEM team 2011]] | ||
| + | |||
| + | |||
| + | See our "Bricks of Knowledge" [https://www.youtube.com/watch?v=7EiVttJpXH4 video on USER cloning] and the [https://2011.igem.org/Team:DTU-Denmark-2 wiki of the 2011 DTU-2 iGEM team.] for more information. | ||
==References== | ==References== | ||
| - | |||
| - | + | [1] Sutton, Mark A., et al. "Too much of a good thing." Nature 472.7342 (2011): 159-161. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [2] Sutton, Mark A., Clare M. Howard, and Jan Willem Erisman, eds. The European nitrogen assessment: sources, effects and policy perspectives. Cambridge University Press, 2011. | |
| - | </ | + | [3] Yaniv D. Scherson, George F. Wells, Sung-Geun Woo, Jangho Lee, Joonhong Park, Brian J. Cantwell and Craig S. Criddle; Nitrogen removal with energy recovery through N<sub>2</sub>O decomposition; Energy & Environmental Science. Issue 1 2013. |
| - | + | [4] Nour-Eldin, HH et al (2010). USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods Mol Biol. 2010;643:185-200. doi: 10.1007/978-1-60761-723-5_13. | |
| + | [5] Nørholm, Morten HH. "A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering." BMC biotechnology 10.1 (2010): 21. | ||
| + | |||
| + | [6] Frandsen, Rasmus, et al. "Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi." BMC molecular biology 9.1 (2008): 70. | ||
| + | |||
| + | |||
| + | U.S. Energy Information Administration, Frequently Asked Questions: How much electricity does an American home use? | ||
| + | |||
| + | Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M.; KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 40, D109-D114 (2012). | ||
| + | |||
| + | Kanehisa, M. and Goto, S.; KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27-30 (2000). | ||
| + | |||
| + | Zumft, Walter G.; Cell Biology and Molecular Basis of Denitrification. Microbiology and Molecular Biology Reviews, December 1997 p. 533–616 | ||
| + | |||
| + | Ingledew, W.J. and Poole, R.K.; The Respiratory Chains of Escherichia coli. Microbiological Reviews, Sept. 1984, p. 222-271 | ||
| + | |||
| + | |||
| + | ==Project Pages== | ||
| + | <html> | ||
| + | <li><a href="/Team:DTU-Denmark/Parts">Parts</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/Timeline">Timeline</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/Notebook">Notebook</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/pBAD_SPL">pBAD SPL</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/Experiments">Experiments</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/Modeling">Modeling</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/Methods">Protocols</a></li> | ||
| + | <li><a href="/Team:DTU-Denmark/HelloWorld">Hello World Pilot Project</a></li> | ||
| + | </html> | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
</div> | </div> | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 22:04, 4 October 2013
Project Description
What?
Our project removes ammonia (NH3) from waste water, and via two E. coli mutants, turns it into nitrous oxide (N2O).
Why?
Global demand for fixed nitrogen has increased to the point that half the human population now relies on chemical fertilizer to grow their food. While fertilizer is a requirement for modern life, runoff from overfertilized farmland can cause eutrophication. In the presence of abundant ammonia, algae overgrow and consume the much of the available oxygen in the water [1]. This results in decreased biodiversity throughout the watershed. Within Europe, 53% of lakes are eutrophic.
In Europe the annual surplus of fixed nitrogen from crop production and livestock farming is 10 Tg (10 Teragrams = a million tonnes), 6 Tg of which leaches into ground and surface water [2]. If that was to be converted into N2O, it could be decomposed for 4.87 Billion kW h -- equivalent to the annual power consumption of 36,000 Avarage households.
If released into the atmosphere, nitrous oxide is a potent greenhouse gas, and capable of degrading ozone, which is why it makes sense to convert it to N2.
How?
Using two E. coli mutants built with genes from Nitrosomonas europaea and Pseudomonas aeruginosa, we provide a system to reverse nitrogen fixation. Our mutants consume ammonia and produce nitrous oxide, releases a sustainable source of energy when decomposed into nitrogen and oxygen [3]. We also provide a model of a bioreactor that could be scaled up and deployed in the field to simultaneously clean the water and produce energy.
Details
Mutant 1: Aerobic
The first mutant incorporates AMO, HAO and two cytochromes c554 and cm552 from Nitrosomonas europaea (shown in blue). AMO is a 3 subunit protein which converts ammonia (NH4) to an intermediate called hydroxylamine (NH2OH). HAO then converts the hydroxylamine to nitrite (NO2-). During this conversion process, cytochrome c554 accepts an electron from HAO, and then passes this electron on to ccm552. The terminal electron acceptor in this chain is quinone. AMO and ccm552 are embedded in the cytoplasmic membrane, and all other proteins in this process are found in the periplasm.
Mutant 2: Anaerobic
The second mutant incorporates the Nir region from Pseudomonas aeruginosa (shown in yellow). NirS converts nitrite to nitric oxide (NO), while removing an electron from NirM. The remainder of the Nir region is necessary for the synthesis of NirS, and so we have included these genes as well. NOR, which is present natively in E. coli, converts nitric oxide to nitrous oxide (N2O). In contrast to N. europaea, E. coli is not a nitrifying bacterium and so does not convert nitrous oxide into nitrogen to complete the denitrification process.Plasmids
Mutant 1
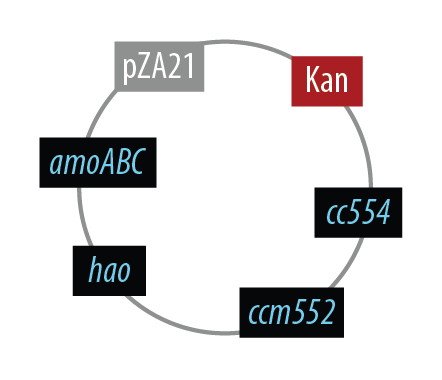
The final plasmid for Mutant 1 should be the following which includes all genes on one plasmid:
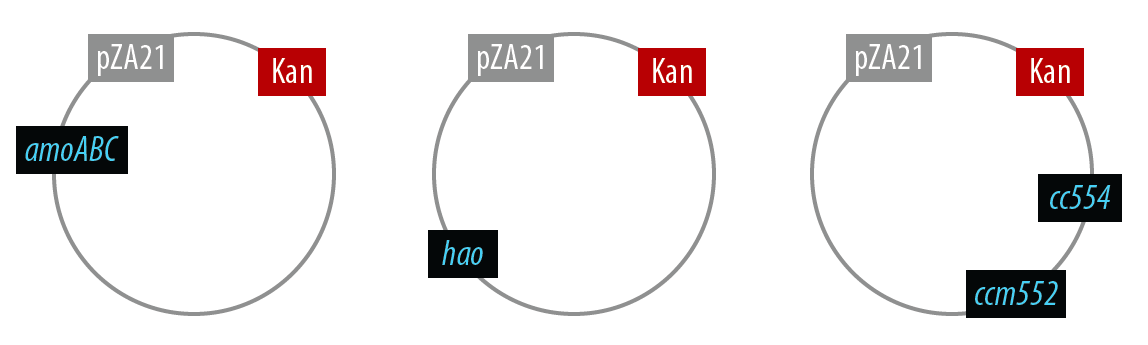
In order to simplify and to be able to debug each part of this pathway separately, we first implemented the genes individually. Unfortunately, we did not have time to combine all the genes into the final plasmid, but we were able to verify that the AMO containing plasmid is working as designed.
Mutant 2
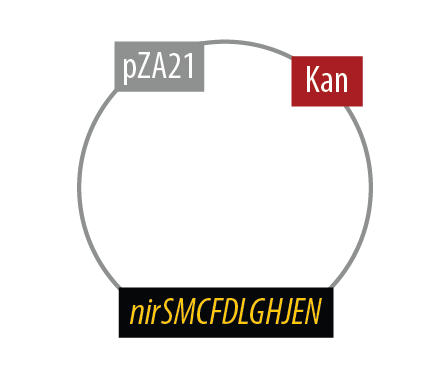
The final plasmid for Mutant 2 is the following:
Since we had difficulty extracting Nir from Pseudomonas aeruginosa in one piece, we split it into two pieces to be extracted seperately and then re-combined them with USER cloning.
USER Cloning
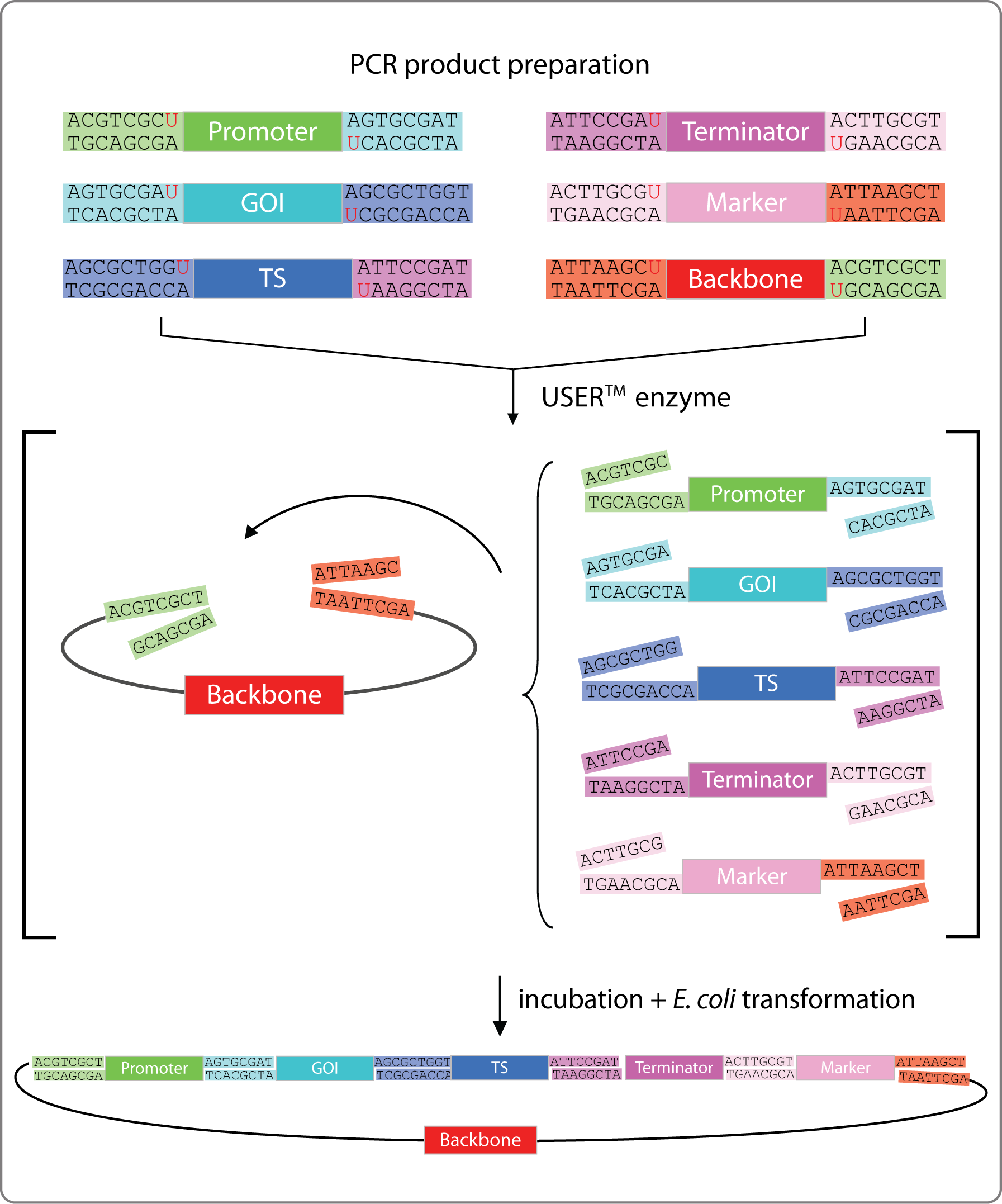
To assemble our plasmids we employ uracil-specific excision reagent (USER) cloning [4]. This technique enables us to speed up the cloning process, to clone seamlessly, and to assemble many fragments in one reaction. The steps are:
- PCR: The backbone and all fragments are amplified as linear products containing a uracil instead of a thymine at a specific position. This requires specially designed set of primers with a tail that will become the overhang and a polymerase that is able to perform uracil-insertion.
- We are using the non-commercial polymerase X7, designed by Morten Nørholm [5].
- Optional - If the method was used to insert genes into a backbone, with it's own resistance marker, the PCR product were treated with DpnI to digest the template DNA from the PCR reaction.
- USER-reaction: Digesting the linearized PCR fragment with Uracil-Specific Excision Reagent (USER) enzyme removes the uracil-insertions. This will produce the required overhangs with the sequence and the length of your wish.
- Transformation: The mix is immediately transformed into competent E. coli cells. The plasmid assembles itself by base pairing between the complementary overhangs and ligation is done by E.coli itself.
Advantages to USER cloning
- You only need one enzyme
- You don't need any ligation reaction
- Seamless assembly
- Directional
- Multiple fragment in one reaction - also in eukaryotes [6].
- Plug 'n' Play
See our "Bricks of Knowledge" video on USER cloning and the wiki of the 2011 DTU-2 iGEM team. for more information.
References
[1] Sutton, Mark A., et al. "Too much of a good thing." Nature 472.7342 (2011): 159-161.
[2] Sutton, Mark A., Clare M. Howard, and Jan Willem Erisman, eds. The European nitrogen assessment: sources, effects and policy perspectives. Cambridge University Press, 2011.
[3] Yaniv D. Scherson, George F. Wells, Sung-Geun Woo, Jangho Lee, Joonhong Park, Brian J. Cantwell and Craig S. Criddle; Nitrogen removal with energy recovery through N2O decomposition; Energy & Environmental Science. Issue 1 2013.
[4] Nour-Eldin, HH et al (2010). USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods Mol Biol. 2010;643:185-200. doi: 10.1007/978-1-60761-723-5_13.
[5] Nørholm, Morten HH. "A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering." BMC biotechnology 10.1 (2010): 21.
[6] Frandsen, Rasmus, et al. "Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi." BMC molecular biology 9.1 (2008): 70.
U.S. Energy Information Administration, Frequently Asked Questions: How much electricity does an American home use?
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M.; KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 40, D109-D114 (2012).
Kanehisa, M. and Goto, S.; KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27-30 (2000).
Zumft, Walter G.; Cell Biology and Molecular Basis of Denitrification. Microbiology and Molecular Biology Reviews, December 1997 p. 533–616
Ingledew, W.J. and Poole, R.K.; The Respiratory Chains of Escherichia coli. Microbiological Reviews, Sept. 1984, p. 222-271
Project Pages
 "
"