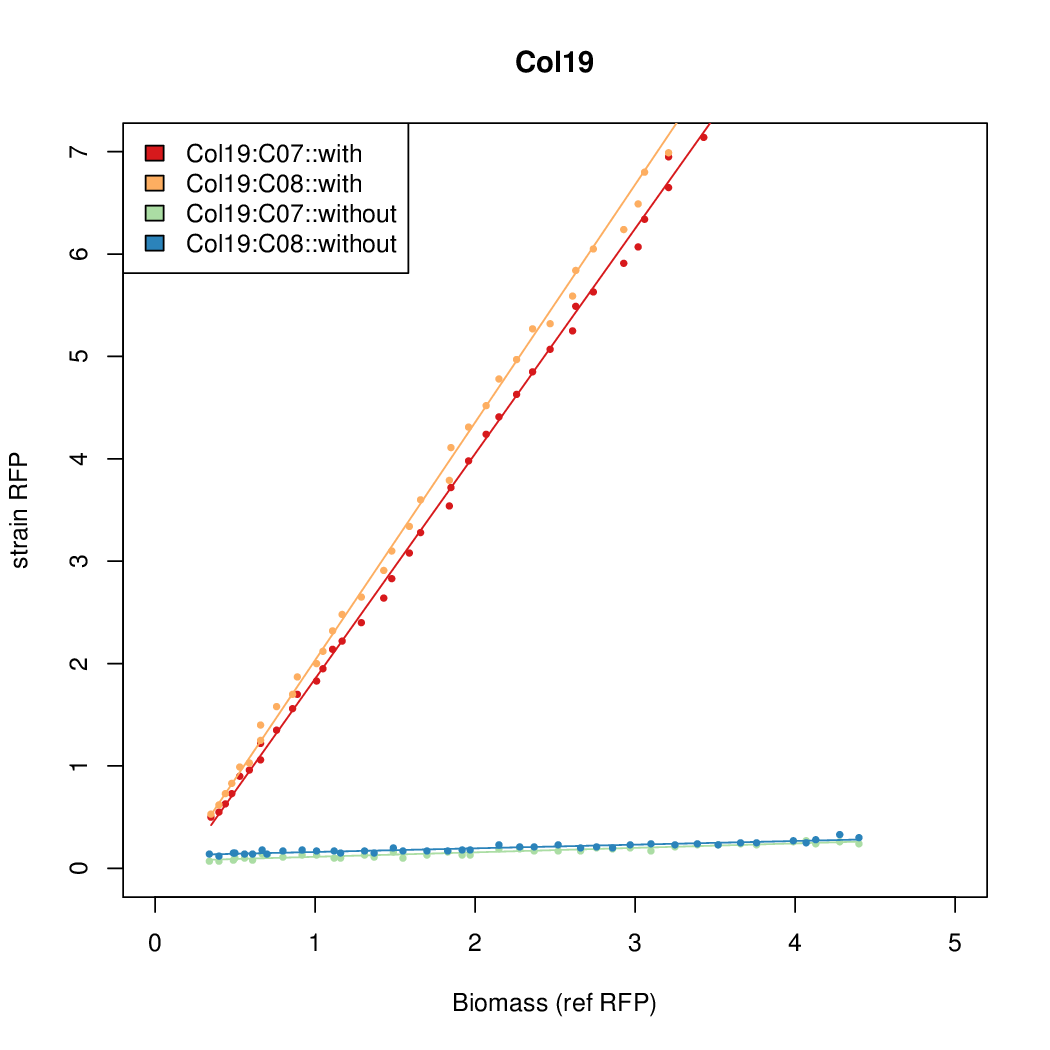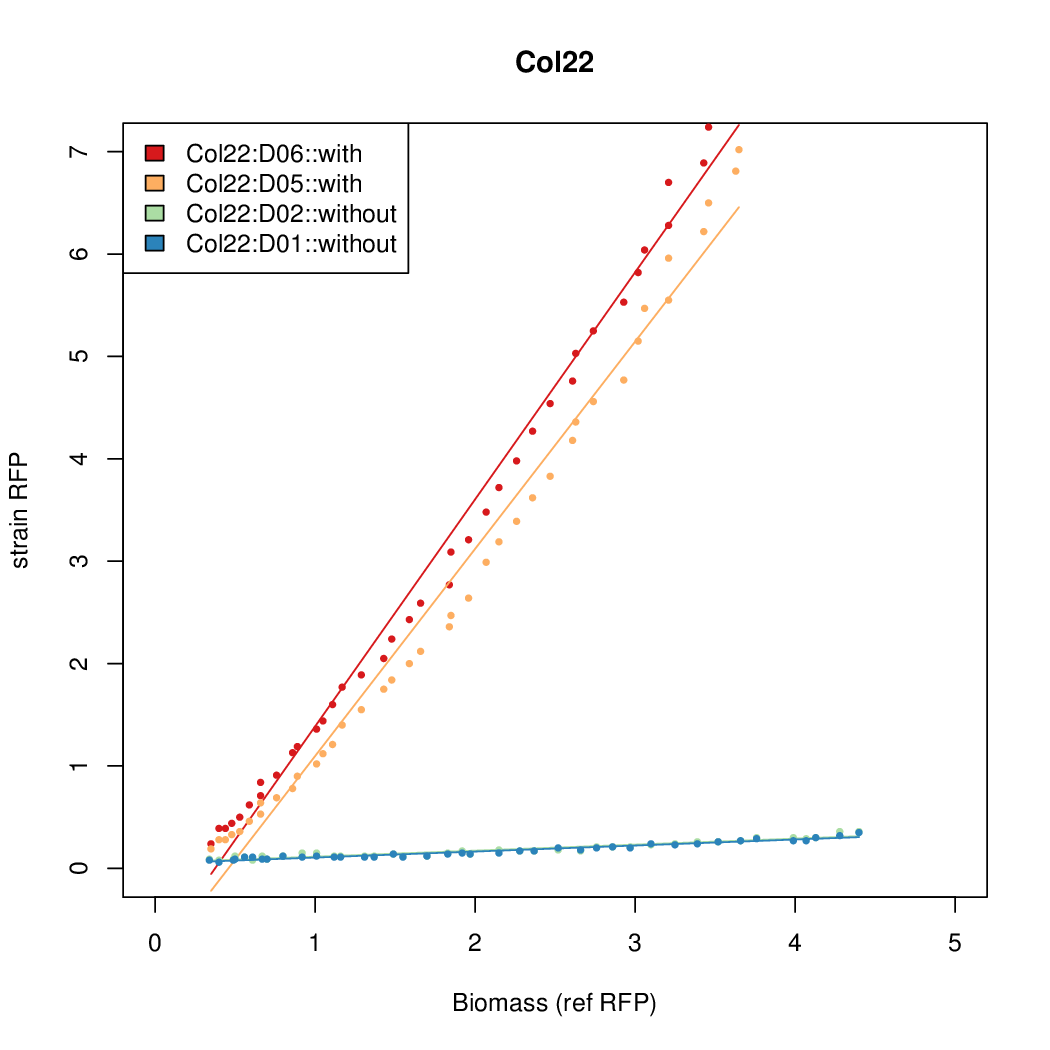Team:DTU-Denmark/pBAD SPL
From 2013.igem.org
(→pBAD synthetic promoter library) |
|||
| Line 5: | Line 5: | ||
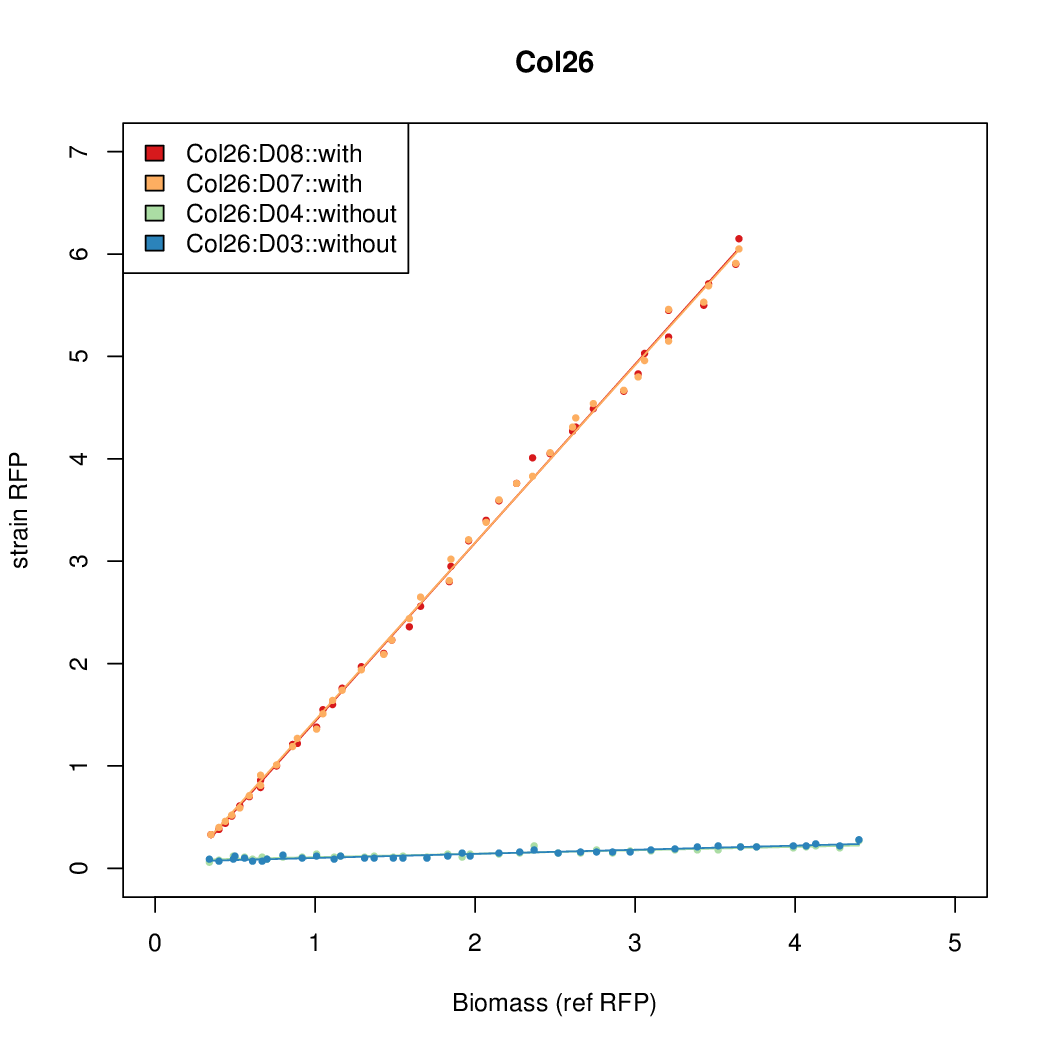
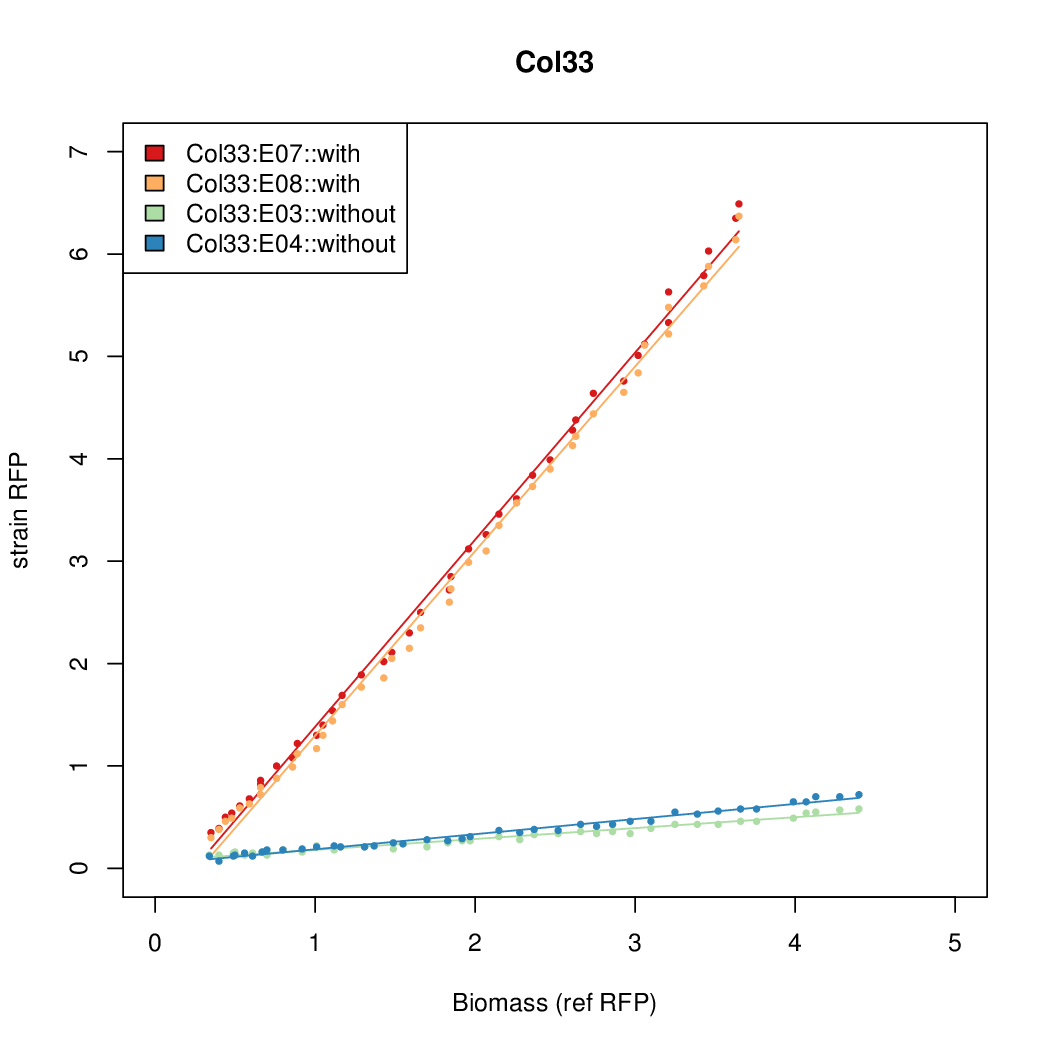
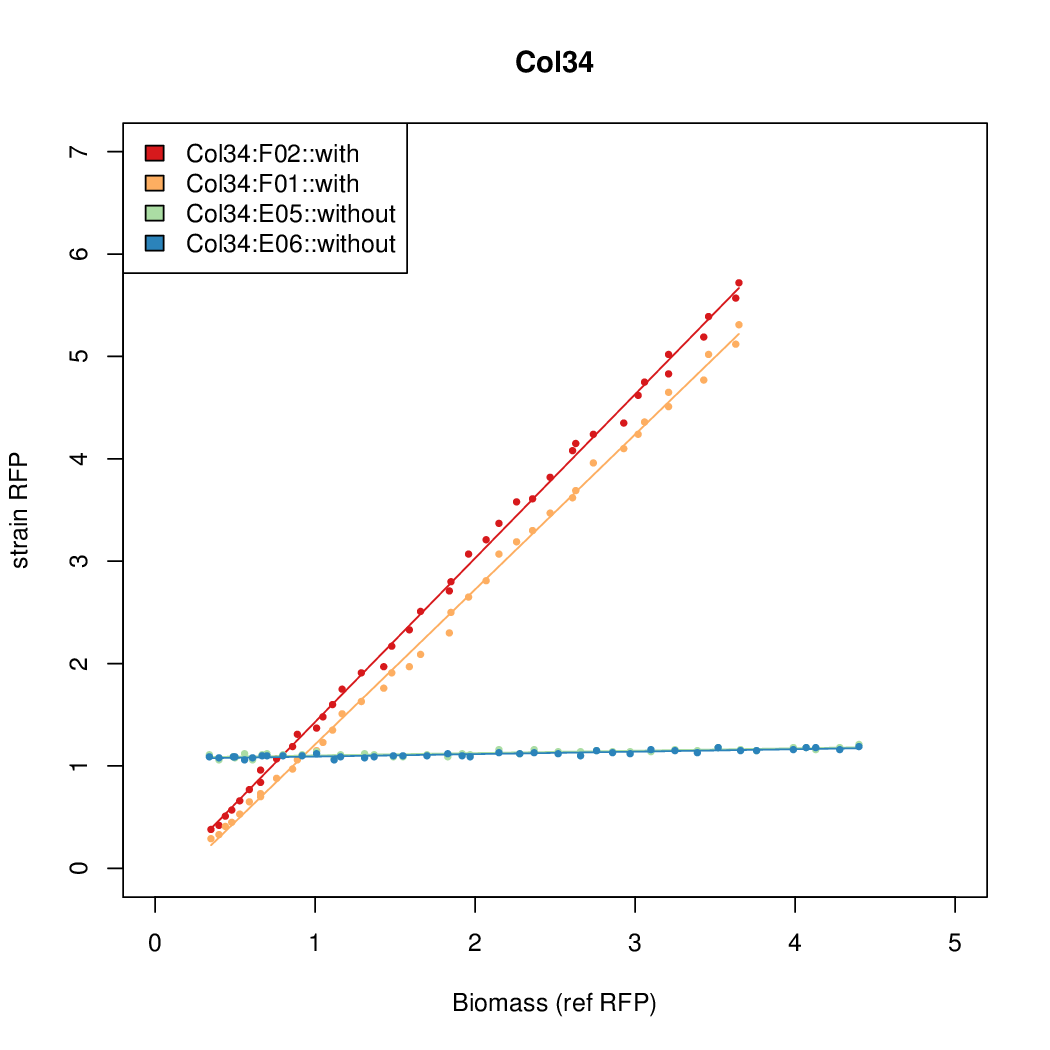
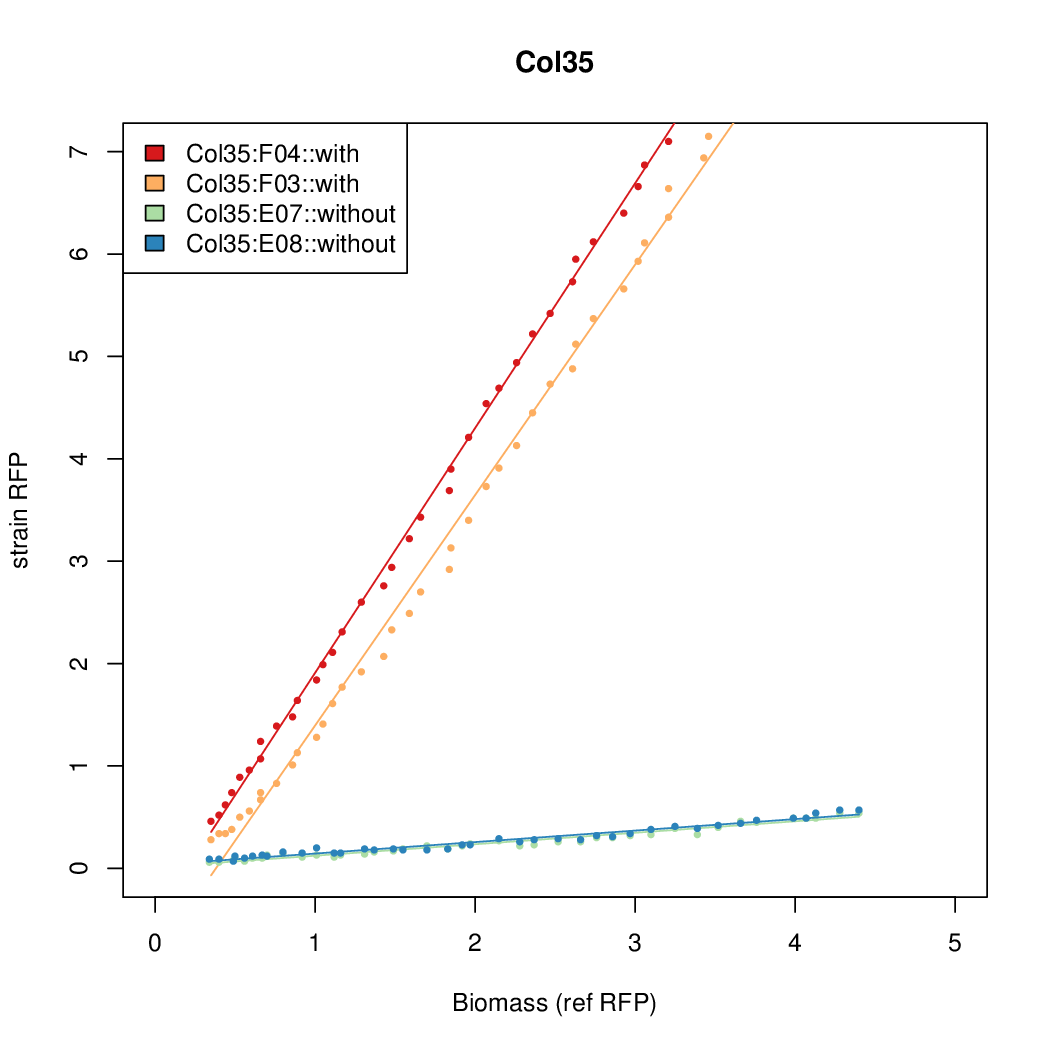
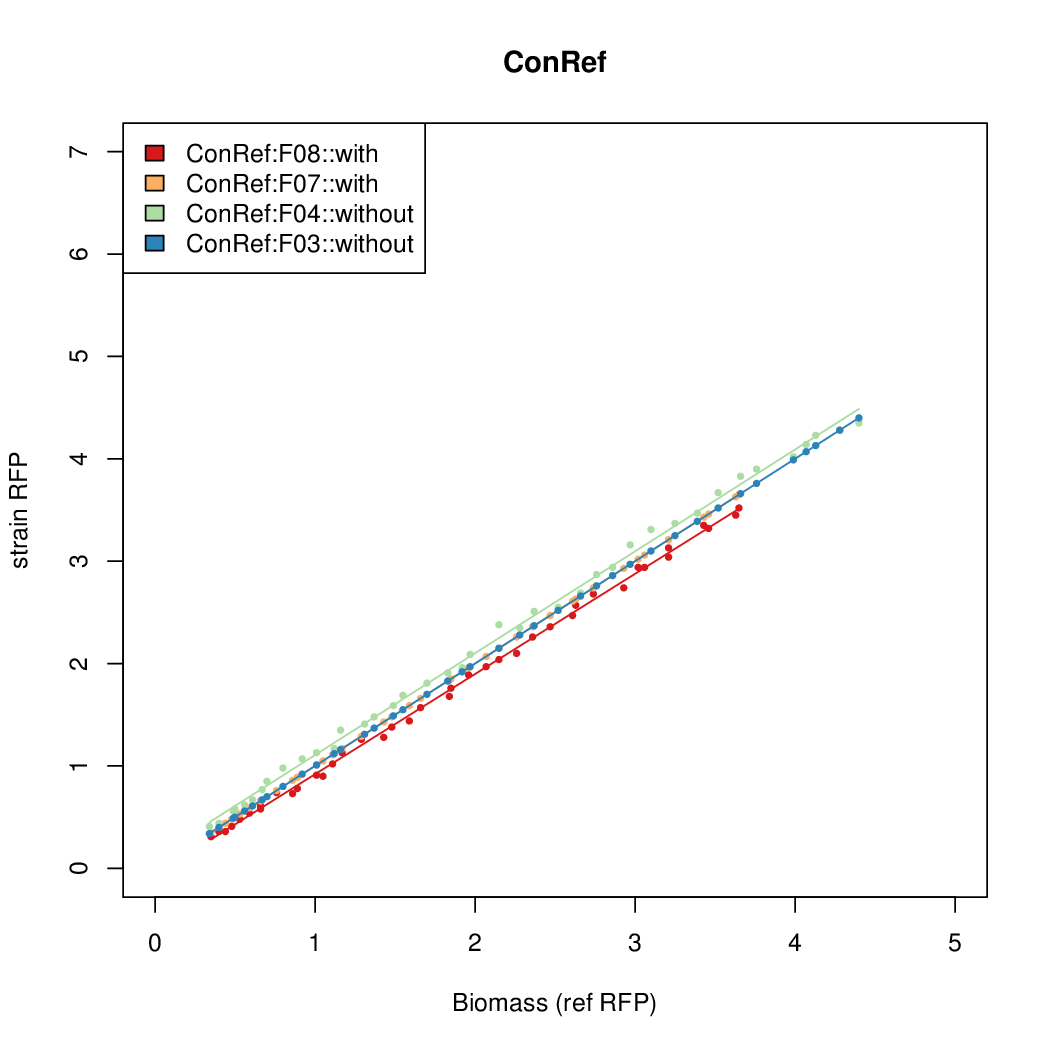
As a tool for expressing lethal proteins in ''E. coli'' we made a synthetic promoter library (SPL, [http://dspace.mit.edu/handle/1721.1/60080 RFC 63]) with the pBAD arabinose inducible promoter. The concept was taken from the [https://2010.igem.org/Team:DTU-Denmark/SPL DTU iGEM team from 2010]. | As a tool for expressing lethal proteins in ''E. coli'' we made a synthetic promoter library (SPL, [http://dspace.mit.edu/handle/1721.1/60080 RFC 63]) with the pBAD arabinose inducible promoter. The concept was taken from the [https://2010.igem.org/Team:DTU-Denmark/SPL DTU iGEM team from 2010]. | ||
| - | == | + | == Methods == |
| + | === Experimental procedure === | ||
| - | + | # Random promoter sequences were ordered matching the sequence CTGACGNNNNNNNNNNNNNNNNNNTAWWATNNNNA. | |
| + | # USER cloning to add RFP downstream of promoter. | ||
| + | # Colonies were plated. | ||
| + | # Colonies that were not red prior to induction with arabinose but that did turn red after induction with arabinose were selected. | ||
| + | # Wells were inoculated from overnight cultures of each of the selected colonies | ||
| + | # One plate was run with arabinose, and another plate was run without arabinose. | ||
| + | === Data analysis === | ||
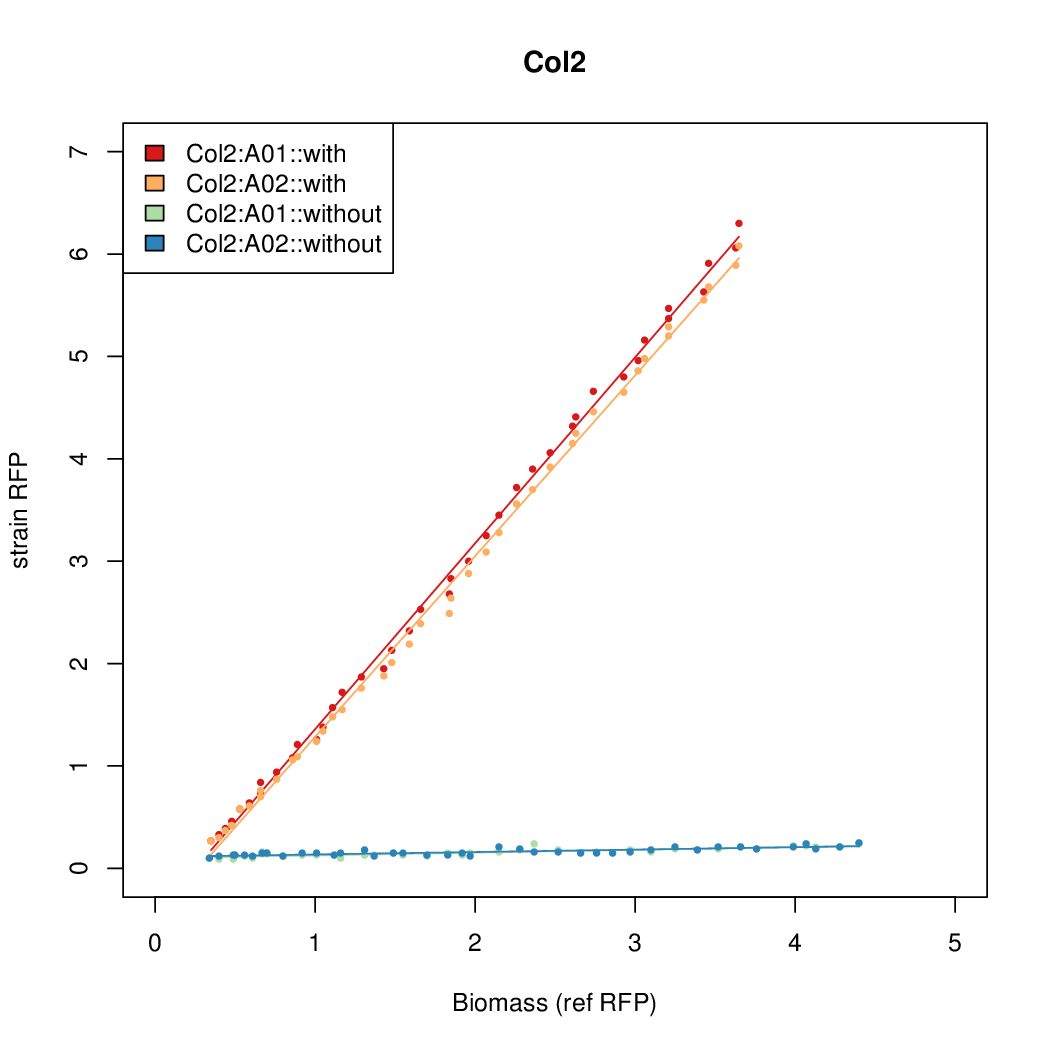
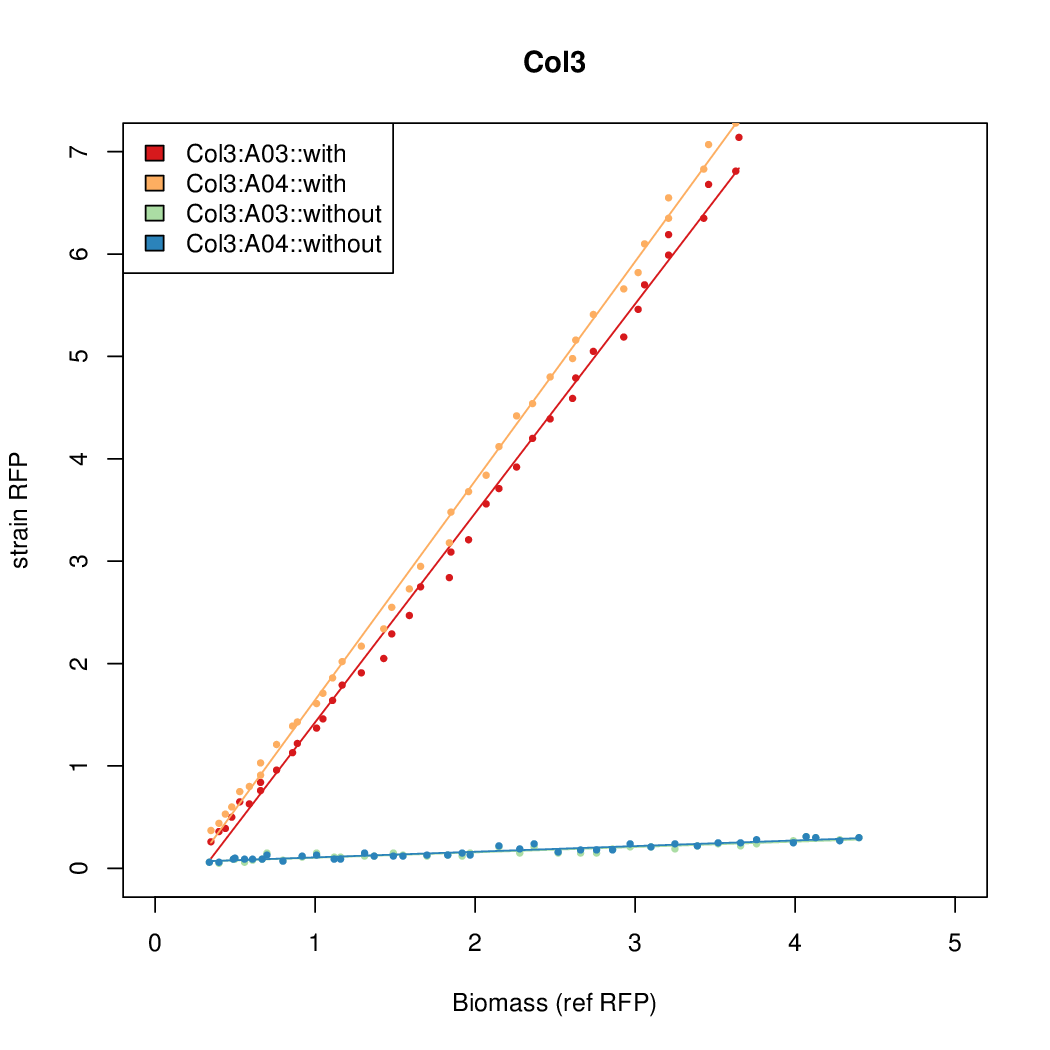
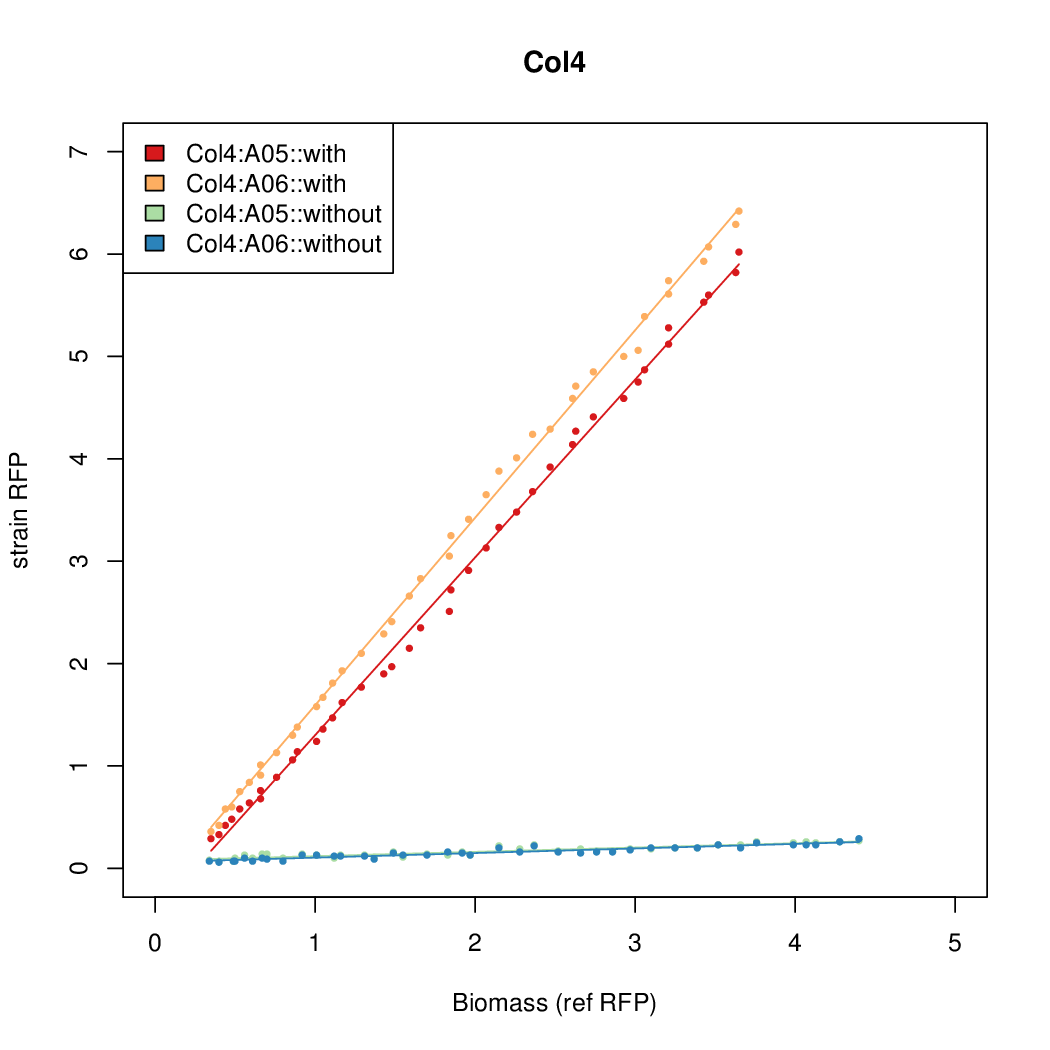
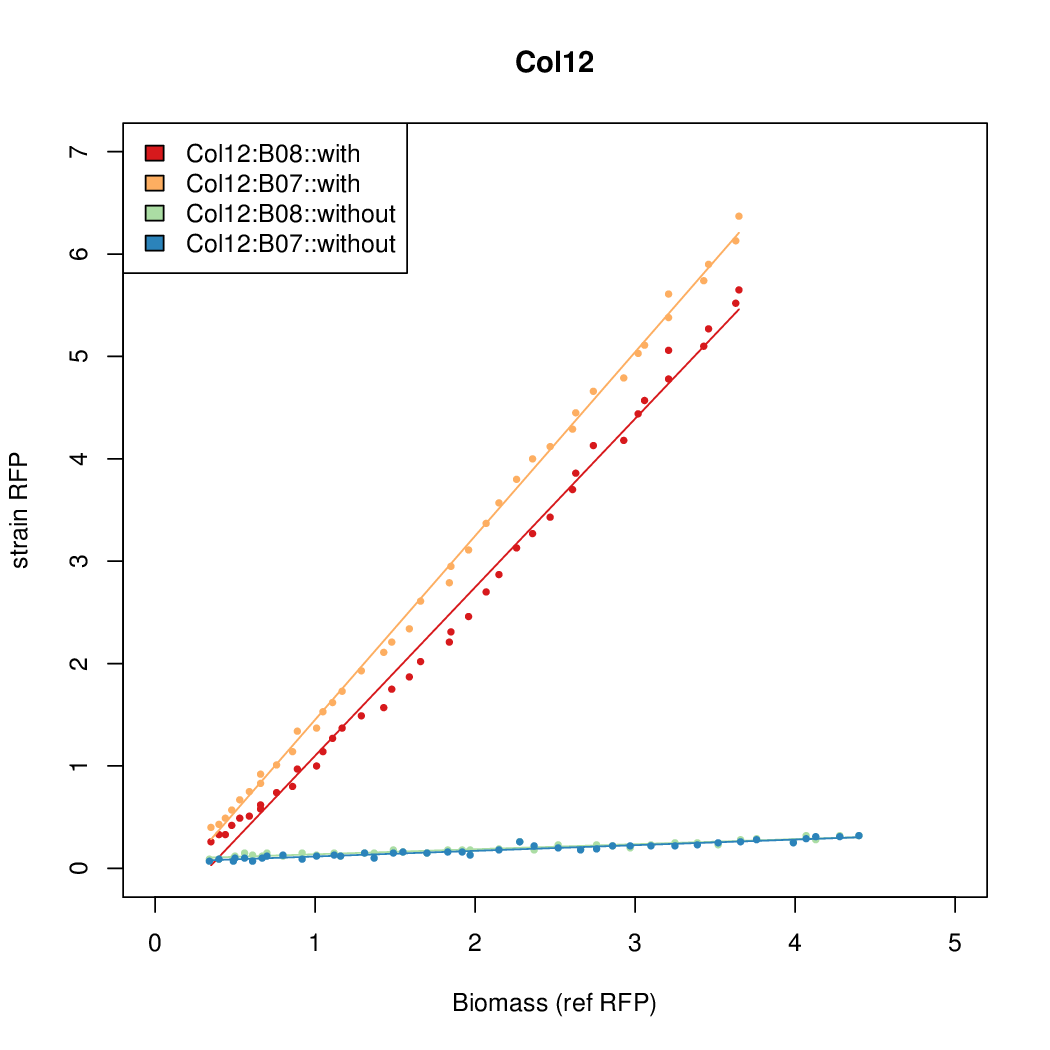
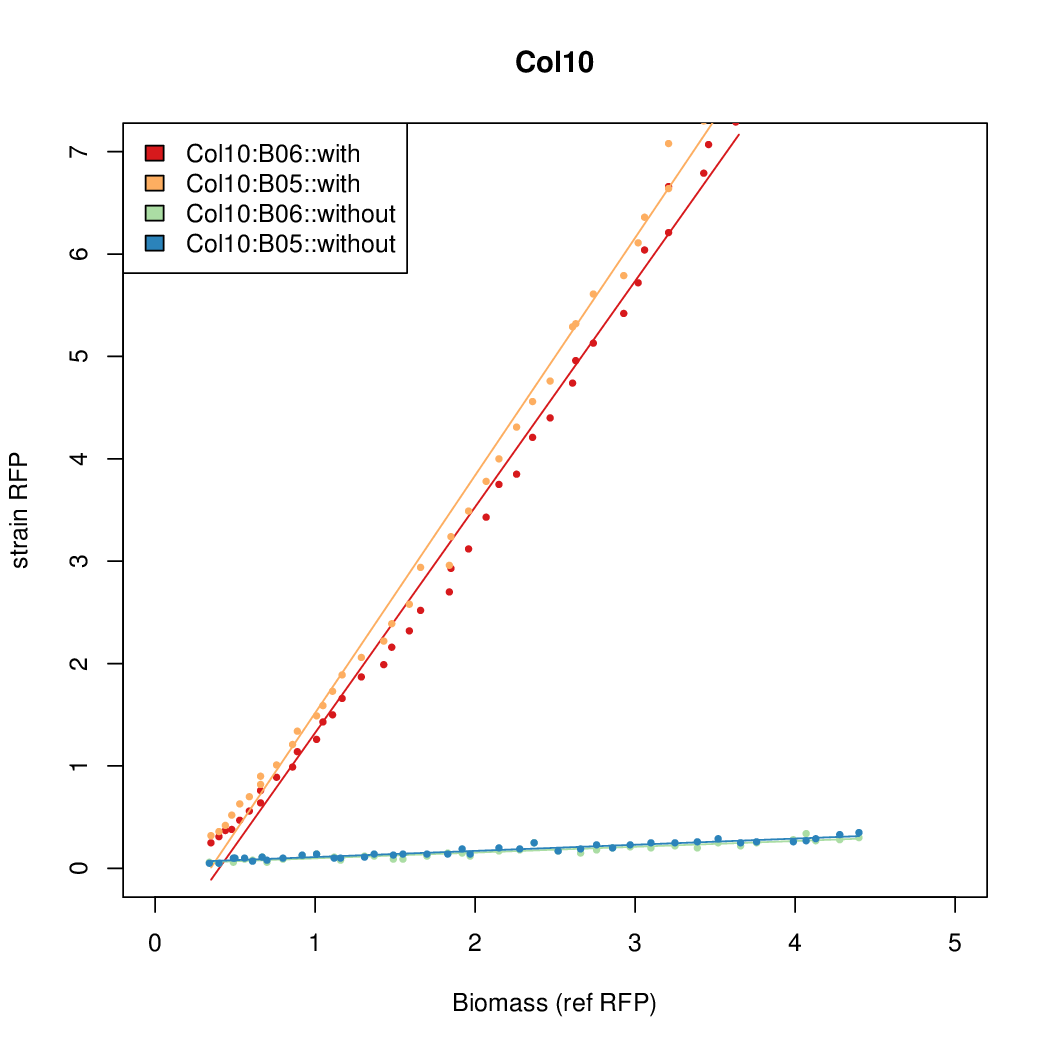
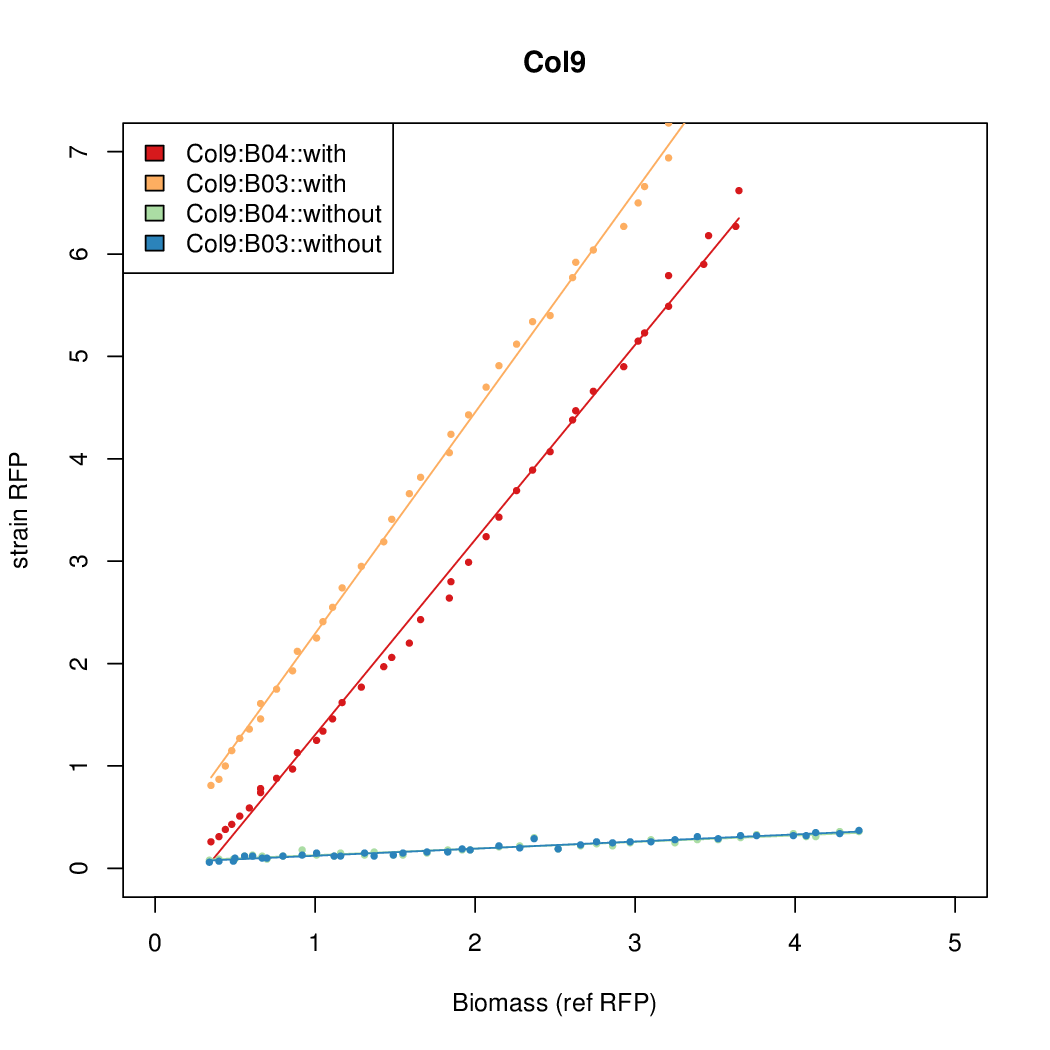
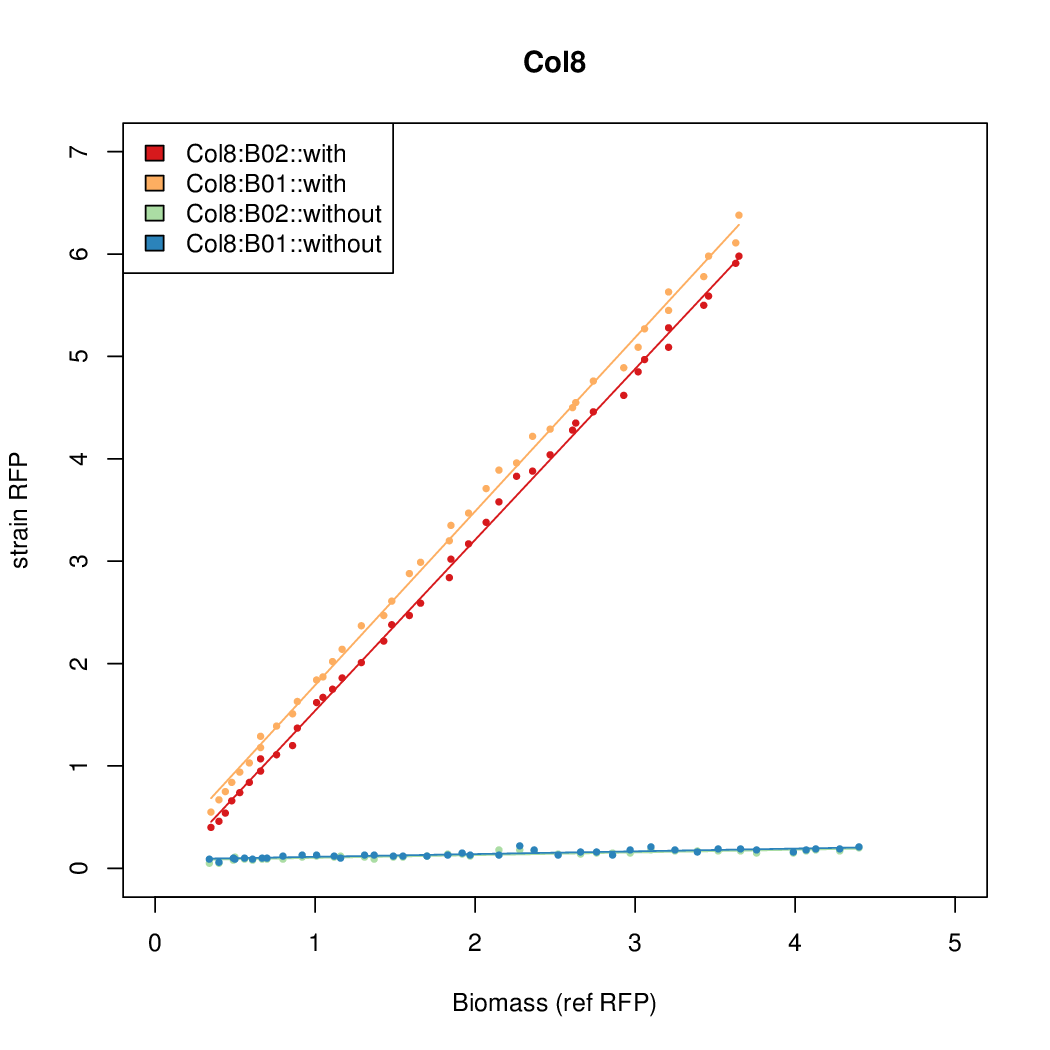
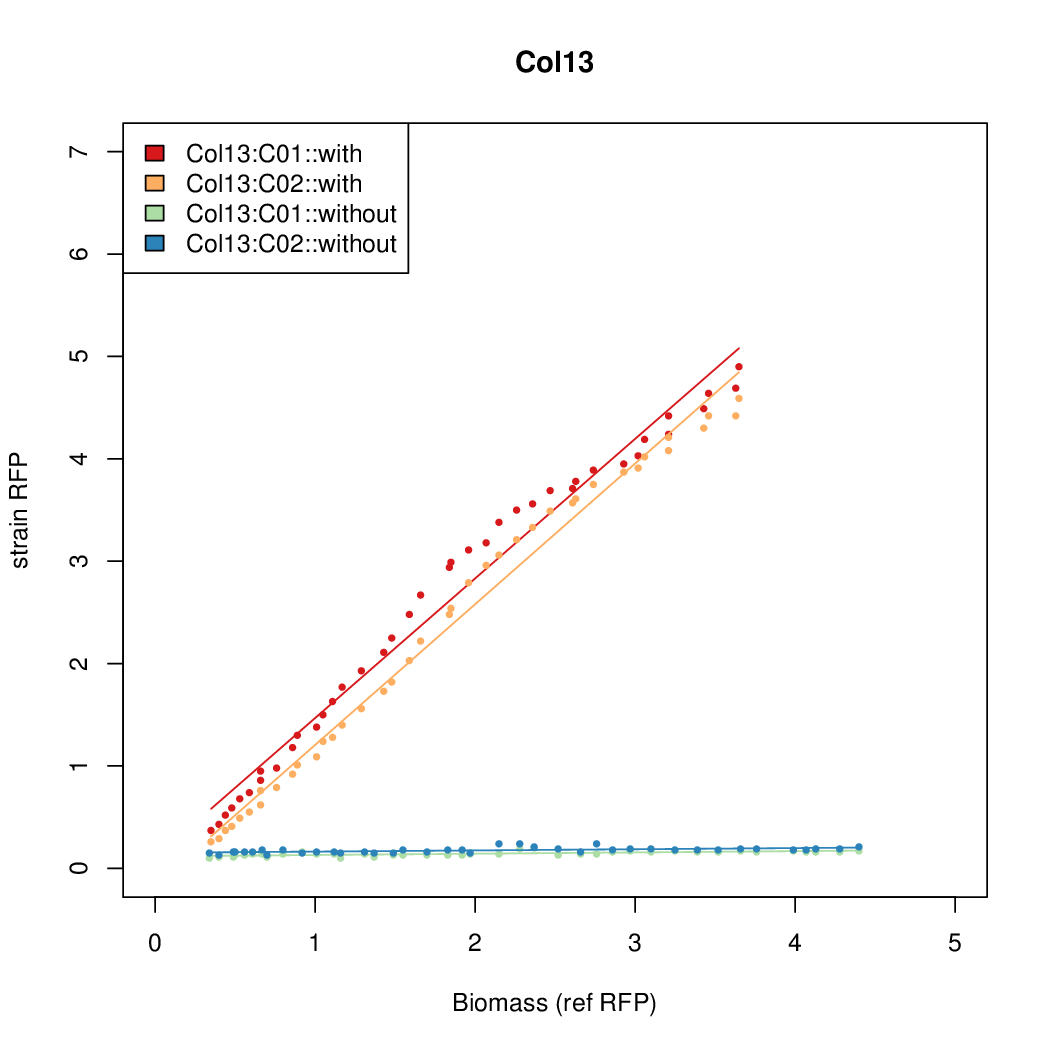
| + | Data was collected from the Biolector, and analyzed using a series of R scripts written by Chris Workman (unpublished). | ||
| + | The growth rate, mu, was estimated to be 1.28 for all wells on all plates since we expect each strain to grow at the same rate. | ||
| + | |||
| + | A time window representing exponential growth was selected (between 1 and 4.5 hours). | ||
Revision as of 07:59, 1 October 2013
pBAD SPL
Contents |
pBAD synthetic promoter library
As a tool for expressing lethal proteins in E. coli we made a synthetic promoter library (SPL, RFC 63) with the pBAD arabinose inducible promoter. The concept was taken from the DTU iGEM team from 2010.
Methods
Experimental procedure
- Random promoter sequences were ordered matching the sequence CTGACGNNNNNNNNNNNNNNNNNNTAWWATNNNNA.
- USER cloning to add RFP downstream of promoter.
- Colonies were plated.
- Colonies that were not red prior to induction with arabinose but that did turn red after induction with arabinose were selected.
- Wells were inoculated from overnight cultures of each of the selected colonies
- One plate was run with arabinose, and another plate was run without arabinose.
Data analysis
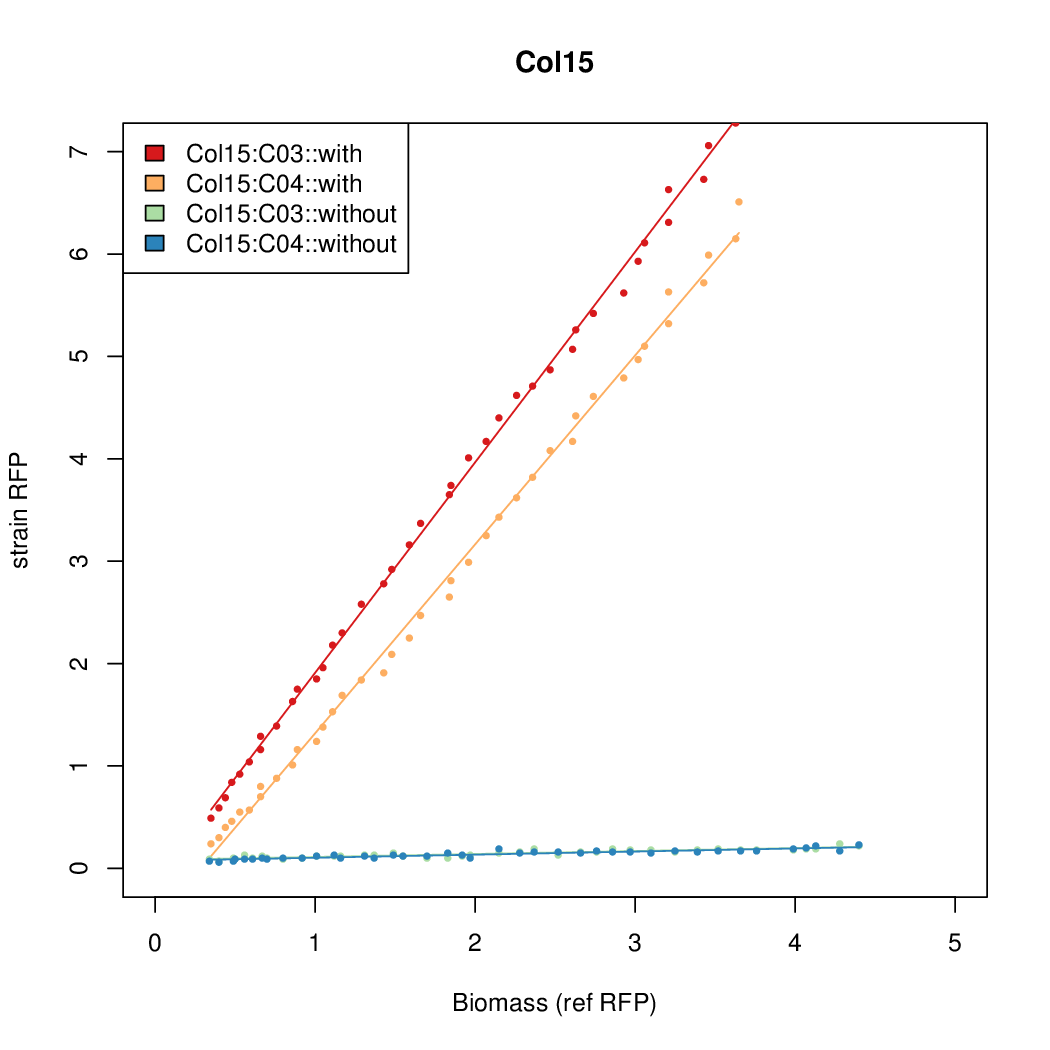
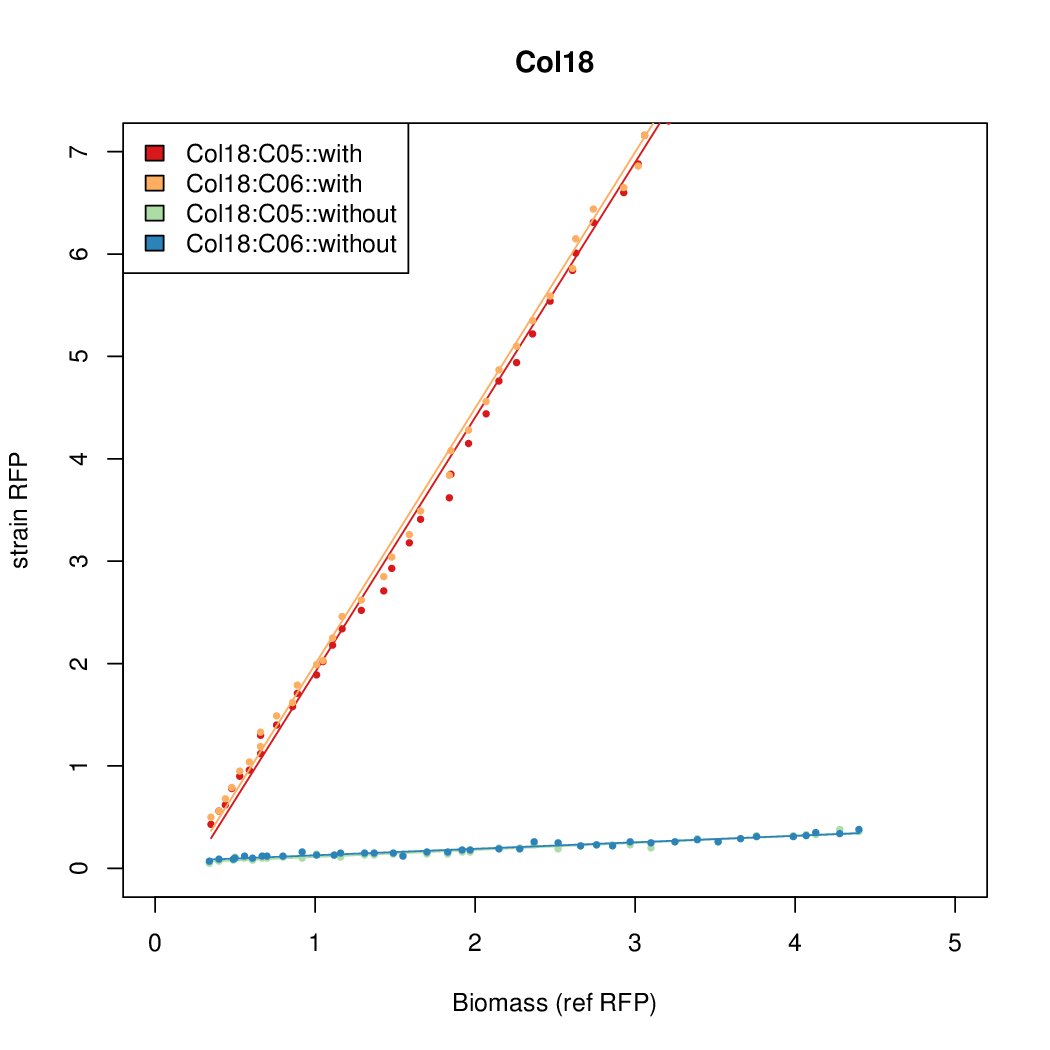
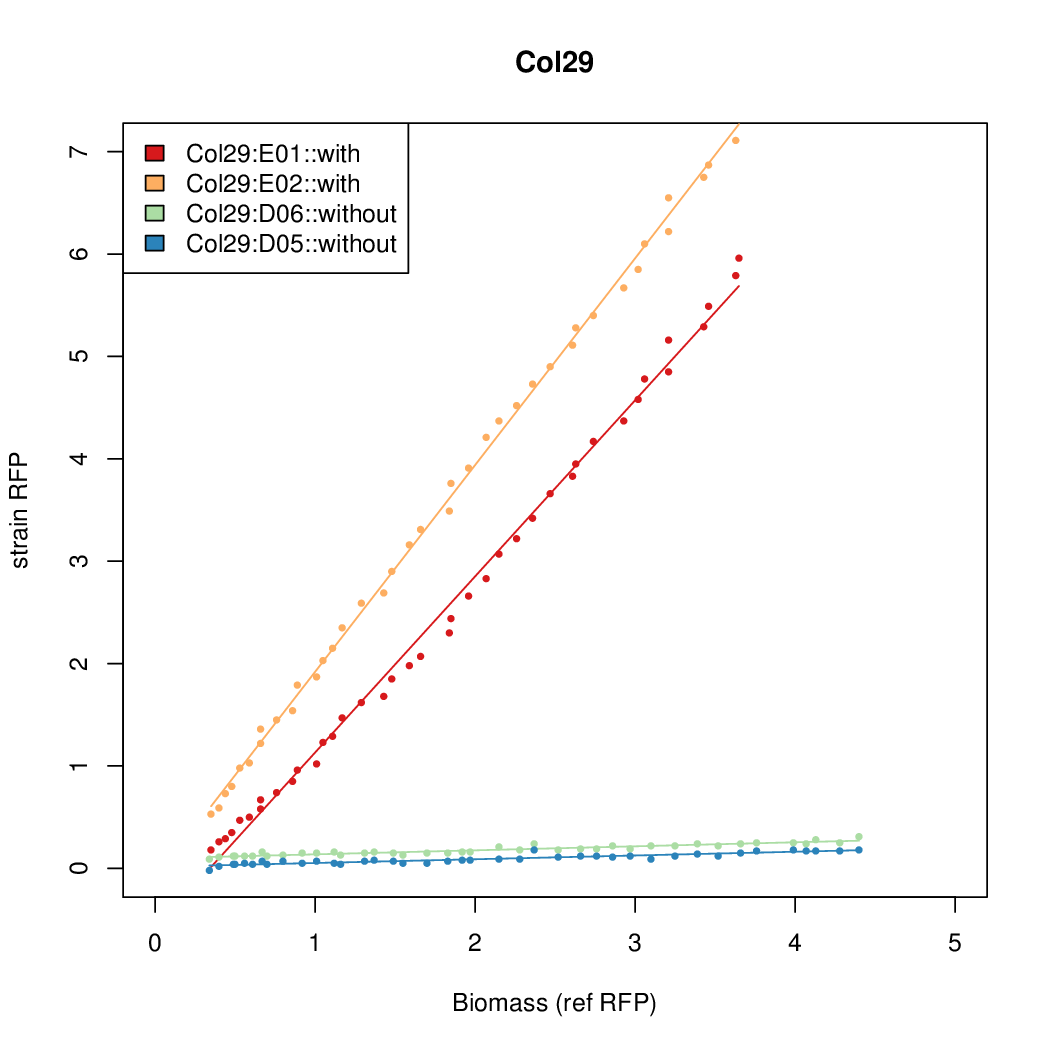
Data was collected from the Biolector, and analyzed using a series of R scripts written by Chris Workman (unpublished).
The growth rate, mu, was estimated to be 1.28 for all wells on all plates since we expect each strain to grow at the same rate.
A time window representing exponential growth was selected (between 1 and 4.5 hours).
Results
Example of use
See also "Hello World project".
 "
"