Team:Dundee/Project/Detector
From 2013.igem.org
(Difference between revisions)
Kyleharrison (Talk | contribs) |
|||
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:Dundee/Templates/Navigationbar}} | ||
| + | |||
<html> | <html> | ||
<html lang="en"> | <html lang="en"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<!-- Begin page content --> | <!-- Begin page content --> | ||
| Line 146: | Line 12: | ||
</div> | </div> | ||
<!-- Title End --> | <!-- Title End --> | ||
| + | |||
| + | <!-- Detector 2 --> | ||
<div class="row" style="text-align:justify;margin-top:-20px;"> | <div class="row" style="text-align:justify;margin-top:-20px;"> | ||
<div class="span12"> | <div class="span12"> | ||
| - | <h2> | + | <h2>The detection systems</h2> |
| - | + | We designed two systems to detect microcystin, one in each of our chassis organisms.<br><br> | |
| - | + | ||
| + | <h2>Engineering the <i>B. subtilis</i> PrkC receptor to respond to microcystin</h2> | ||
| + | <i>B. subtilis</i> forms desiccation-resistant structures called spores in order to survive harsh environmental conditions. In order for spores to know that the conditions have become favourable for germination and growth they must monitor the extracellular environment. This is achieved through a number of inner-membrane receptors described as germinant receptors. PrkC is an example of a germinant receptor and it binds to cell wall-associated peptides.<br><br> | ||
| + | <h2>Why sense cell wall peptides? How does this indicate that conditions are permissive for growth?</h2> | ||
| + | Actively growing cells turnover cell wall components and these are released into the extracellular milieu. So by sensing cell wall components, through the PrkC receptor, the spore can tell that other cells are growing in the nearby environment. This is how the PrkC receptor can signal to the spore that conditions are permissive for growth. PrkC receptor activation triggers a process called germination, which is the conversion of the spore back into an actively growing cell.<br><br> | ||
| - | + | <h2>The PrkC receptor</h2> | |
| + | The extracellular portion of the PrkC receptor has 4 domains. Three of these are PASTA domains which are capped by a forth, C-terminal domain. The protein is anchored in the inner membrane and has an N-terminal kinase domain that phosphorylates downstream targets upon receptor activation. The 3 PASTA domains are implicated in binding of the cell wall components and are thus described as ligand binding domains (Fig 1A).<br><br> | ||
| + | <h2>But how can we use the PrkC receptor to detect microcystin?</h2> | ||
| + | We hope to detect microcystin by replacing the 3 ligand binding domains with three copies of PP1 (Fig 1B).<br><br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <center><img src="http://farm8.staticflickr.com/7292/10034973665_c91f4f9ea7_o.jpg"><br></center> | |
| + | <strong>Figure 1. : The <i>B. subtilis</i> PrkC Receptor (A) and the engineered receptor designed to respond to microcystin (B). </strong><br><br> | ||
| + | We hope that when microcystin binds to the PP1 regions of the modified PrkC receptor this will result in activation of the downstream pathways controlled by native PrkC. Additionally, we aim to have our <i>B. subtilis</i> strain constitutively expressing GFP so that when it is relieved from dormancy it will fluoresce and this will be detectable with our electronic <a href="https://2013.igem.org/Team:Dundee/Project/SoftwareTheory" target="_blank">Moptopus device</a>.<br><br> | ||
| + | <h2>Progress so far</h2> | ||
| + | We are currently in the process of cloning this receptor and we are having some difficulty. We have successfully cloned the 5’ part of <i>prkC</i>, encoding the kinase domain, and are currently in the process of sequentially adding the PP1 genes by suicide ligation. The final step after this will be to ligate the 3’ end of <i>prkC</i>.<br><br> | ||
| - | + | <h2>Engineering the <i>E. coli</i> EnvZ sensor kinase to respond to microcystin</h2> | |
| - | + | The EnvZ system is a signal transduction system composed of two parts and is, therefore, described as a two-component regulatory system. Part 1 is the sensor kinase protein located in the cell envelope and Part 2 is the cytoplasmic response regulator protein. The native EnvZ sensor detects changes in osmolarity.<br><br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <h2>EnvZ sensor kinase</h2> | |
| + | The sensor kinase EnvZ detects a signal from the environment and auto-phosphorylates. The phosphoryl group is then transferred to the response regulator OmpR. OmpR is a DNA-binding protein. <i>E. coli</i> is a Gram-negative bacterium which means that it has inner and outer membranes. The EnvZ sensor sits on the inner membrane (Fig 2).<br><br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <center><img src="http://farm3.staticflickr.com/2859/10035077703_80c60c236e_o.jpg"></center><br> | |
| - | < | + | |
| + | <strong>Figure 2. The Native <i>E. coli</i> EnvZ Receptor.</strong> The N- and C-termini of EnvZ are located in the cytoplasm, with two transmembrane domains separated by a periplasmic loop. The periplasmic loop senses membrane tension caused by osmotic stress. This tension is transmitted the cytoplasmic side of the protein and triggers auto-phosphorylation.<br><br> | ||
| + | <h2>EnvZ sensor to detect microcystin</h2> | ||
| + | We want to replace the periplasmic domain of EnvZ with the PP1 protein (Fig 3), so that when microcystin binds to PP1 it will activate the receptor. This will lead to the phosphorylation and activation of the DNA binding protein OmpR. We will also express in our engineered bacteria a DNA construct encoding the GFP gene under control of the ompC promoter. This promoter is recognised and activated by phosphorylated OmpR and as a result, cells will turn green in the presence of microcystin, this acting as a microcystin detector.<br><br> | ||
| - | < | + | <center><img src="http://farm8.staticflickr.com/7415/10035089443_977abb0772_o.jpg"></center><br> |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <strong>Figure 3. Schematic representation of the engineered EnvZ microcystin detector.</strong> In the engineered construct PP1 replaces the periplasmic domain of EnvZ.<br><br> | ||
| + | <h2>Progress</h2> | ||
| - | + | So far we have successfully cloned the 5’ and 3’ parts of </i>envZ</i>, replacing DNA encoding the periplasmic loop with that of the PP1 gene. Although this construct has been verified by sequencing, to date our attempts to express this hybrid protein have been unsuccessful. A possible reason for this is that the hybrid receptor is not correctly assembled. The periplasmic part of the receptor is translocated across the membrane by the Sec pathway, and we have already seen in our <a href="https://2013.igem.org/Team:Dundee/Project/MopMaking" target="_blank">mop experiments</a> that PP1 cannot be transported by Sec (possibly due to the presence of 13 cysteine residues in PP1 that may be aberrantly disulphide-bonded after translocation). It may be possible to overcome these limitations by re-engineering our hybrid EnvZ to interact with the Tat rather than the Sec pathway. | |
| - | + | The reporter under the control of OmpR has successfully been constructed, and <a href="https://2013.igem.org/Team:Dundee/Project/ReporterOmpC" target="_blank">confirmed to respond to OmpR by a change in expression of GFP.</a><br><br> | |
| + | <h2>Characterisation of our receptors</h2> | ||
| + | We ultimately want to quantify how many of our PrkC receptors are expressed on the surface of the spores and also how many EnvZ sensors are present on our <i>E. coli</i> cells.<br><br> | ||
| - | + | We have purchased microcystin to test our mop, and we can use this to bind and activate our receptors. We will then measure the amount of fluorescence by flow cytometry or microscopy. Then we can quantify the expression of GFP in relation to how much microcystin is presented to our cells. Using the number of receptors expressed in the membrane/spore for each cell we can calculate the effectiveness of our detector. | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
Latest revision as of 10:56, 2 October 2013
The Detector
The detection systems
We designed two systems to detect microcystin, one in each of our chassis organisms.Engineering the B. subtilis PrkC receptor to respond to microcystin
B. subtilis forms desiccation-resistant structures called spores in order to survive harsh environmental conditions. In order for spores to know that the conditions have become favourable for germination and growth they must monitor the extracellular environment. This is achieved through a number of inner-membrane receptors described as germinant receptors. PrkC is an example of a germinant receptor and it binds to cell wall-associated peptides.Why sense cell wall peptides? How does this indicate that conditions are permissive for growth?
Actively growing cells turnover cell wall components and these are released into the extracellular milieu. So by sensing cell wall components, through the PrkC receptor, the spore can tell that other cells are growing in the nearby environment. This is how the PrkC receptor can signal to the spore that conditions are permissive for growth. PrkC receptor activation triggers a process called germination, which is the conversion of the spore back into an actively growing cell.The PrkC receptor
The extracellular portion of the PrkC receptor has 4 domains. Three of these are PASTA domains which are capped by a forth, C-terminal domain. The protein is anchored in the inner membrane and has an N-terminal kinase domain that phosphorylates downstream targets upon receptor activation. The 3 PASTA domains are implicated in binding of the cell wall components and are thus described as ligand binding domains (Fig 1A).But how can we use the PrkC receptor to detect microcystin?
We hope to detect microcystin by replacing the 3 ligand binding domains with three copies of PP1 (Fig 1B).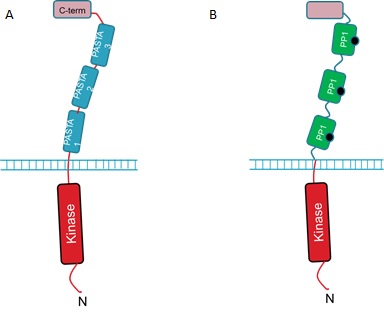
We hope that when microcystin binds to the PP1 regions of the modified PrkC receptor this will result in activation of the downstream pathways controlled by native PrkC. Additionally, we aim to have our B. subtilis strain constitutively expressing GFP so that when it is relieved from dormancy it will fluoresce and this will be detectable with our electronic Moptopus device.
Progress so far
We are currently in the process of cloning this receptor and we are having some difficulty. We have successfully cloned the 5’ part of prkC, encoding the kinase domain, and are currently in the process of sequentially adding the PP1 genes by suicide ligation. The final step after this will be to ligate the 3’ end of prkC.Engineering the E. coli EnvZ sensor kinase to respond to microcystin
The EnvZ system is a signal transduction system composed of two parts and is, therefore, described as a two-component regulatory system. Part 1 is the sensor kinase protein located in the cell envelope and Part 2 is the cytoplasmic response regulator protein. The native EnvZ sensor detects changes in osmolarity.EnvZ sensor kinase
The sensor kinase EnvZ detects a signal from the environment and auto-phosphorylates. The phosphoryl group is then transferred to the response regulator OmpR. OmpR is a DNA-binding protein. E. coli is a Gram-negative bacterium which means that it has inner and outer membranes. The EnvZ sensor sits on the inner membrane (Fig 2).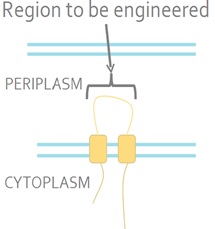
Figure 2. The Native E. coli EnvZ Receptor. The N- and C-termini of EnvZ are located in the cytoplasm, with two transmembrane domains separated by a periplasmic loop. The periplasmic loop senses membrane tension caused by osmotic stress. This tension is transmitted the cytoplasmic side of the protein and triggers auto-phosphorylation.
EnvZ sensor to detect microcystin
We want to replace the periplasmic domain of EnvZ with the PP1 protein (Fig 3), so that when microcystin binds to PP1 it will activate the receptor. This will lead to the phosphorylation and activation of the DNA binding protein OmpR. We will also express in our engineered bacteria a DNA construct encoding the GFP gene under control of the ompC promoter. This promoter is recognised and activated by phosphorylated OmpR and as a result, cells will turn green in the presence of microcystin, this acting as a microcystin detector.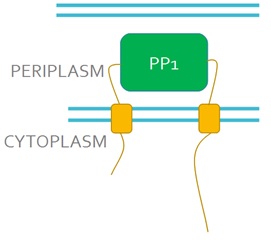
Figure 3. Schematic representation of the engineered EnvZ microcystin detector. In the engineered construct PP1 replaces the periplasmic domain of EnvZ.
Progress
So far we have successfully cloned the 5’ and 3’ parts of envZ, replacing DNA encoding the periplasmic loop with that of the PP1 gene. Although this construct has been verified by sequencing, to date our attempts to express this hybrid protein have been unsuccessful. A possible reason for this is that the hybrid receptor is not correctly assembled. The periplasmic part of the receptor is translocated across the membrane by the Sec pathway, and we have already seen in our mop experiments that PP1 cannot be transported by Sec (possibly due to the presence of 13 cysteine residues in PP1 that may be aberrantly disulphide-bonded after translocation). It may be possible to overcome these limitations by re-engineering our hybrid EnvZ to interact with the Tat rather than the Sec pathway. The reporter under the control of OmpR has successfully been constructed, and confirmed to respond to OmpR by a change in expression of GFP.Characterisation of our receptors
We ultimately want to quantify how many of our PrkC receptors are expressed on the surface of the spores and also how many EnvZ sensors are present on our E. coli cells.We have purchased microcystin to test our mop, and we can use this to bind and activate our receptors. We will then measure the amount of fluorescence by flow cytometry or microscopy. Then we can quantify the expression of GFP in relation to how much microcystin is presented to our cells. Using the number of receptors expressed in the membrane/spore for each cell we can calculate the effectiveness of our detector.
 "
"
