Team:Greensboro-Austin/MAPs
From 2013.igem.org
Mhammerling (Talk | contribs) (→Non-canonical Amino Acids) |
|||
| Line 27: | Line 27: | ||
==Non-canonical Amino Acids== | ==Non-canonical Amino Acids== | ||
| - | The "genetic code" refers to | + | The "genetic code" refers to the rules by which RNA is translated into the proteins which perform most cellular functions. The genetic code is highly conserved throughout all organisms on earth, with 20 amino acids considered to be "canonical". However, many organisms encode non-canonical amino acids, or will modify residues after translation. Therefore, it is possible to transfer these exceptions to the code into organisms commonly used in molecular biology such as ''E. coli''. This increases the toolbox of reactions and functions available when engineering new proteins. |
One such recoding method is repurposing one of the stop codons in ''E. coli''. There are three stop codons available, and in ''E. coli'' the Amber stop codon, UAG, is used relatively rarely, and therefore presents an excellent target for recoding. In fact, this recoding already occurs in the Archaea ''Methanocaldococcus jannaschii''. ''M. jannashii'' has a tyrosyl-tRNA synthetase pair where the tRNA is charged with tyrosine and the anticodon loop recognizes the Amber stop codon. The active site of the orthogonal synthetase can be mutated to charge the tRNA with a non-canonical amino acid instead of tyrosine. [[#References|[1]]] | One such recoding method is repurposing one of the stop codons in ''E. coli''. There are three stop codons available, and in ''E. coli'' the Amber stop codon, UAG, is used relatively rarely, and therefore presents an excellent target for recoding. In fact, this recoding already occurs in the Archaea ''Methanocaldococcus jannaschii''. ''M. jannashii'' has a tyrosyl-tRNA synthetase pair where the tRNA is charged with tyrosine and the anticodon loop recognizes the Amber stop codon. The active site of the orthogonal synthetase can be mutated to charge the tRNA with a non-canonical amino acid instead of tyrosine. [[#References|[1]]] | ||
| Line 33: | Line 33: | ||
| - | Attempting to recode the Amber codon leads to several complications. There are several essential genes which terminate in the Amber stop codon, and which do not function properly if a non-canonical amino acid is used instead of translation stopping. Additionally, the release factor RF1 is responsible for terminating translation when the ribosome reaches an Amber stop codon, and will compete with the tRNA carrying the non-canonical amino acid. In order to fully recode the Amber codon, the strain RF0 [[#References|[2]]] | + | Attempting to recode the Amber codon leads to several complications. There are several essential genes which terminate in the Amber stop codon, and which do not function properly if a non-canonical amino acid is used instead of translation stopping. Additionally, the release factor RF1 is responsible for terminating translation when the ribosome reaches an Amber stop codon, and will compete with the tRNA carrying the non-canonical amino acid. In order to fully recode the Amber codon, the strain RF0 [[#References|[2]]] contains an F plasmid with all the essential genes normally ending in the amber codon mutated to end in another stop codon, and has RF1 knocked out to facilitate ncAA incorporation. |
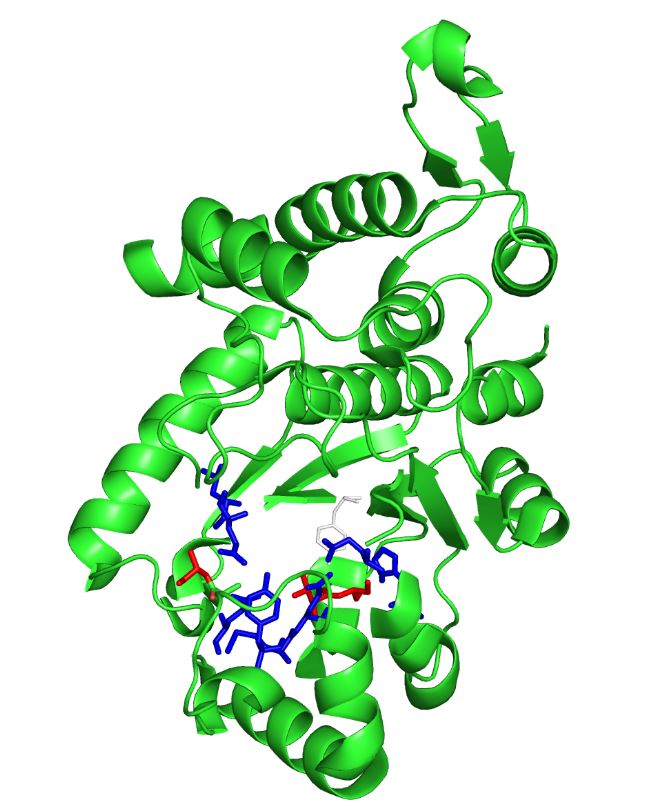
| - | The image on the left is the ''M. jannaschii'' synthetase mutated to incorporate | + | The image on the left is the ''M. jannaschii'' synthetase mutated to incorporate 3-iodotyrosine by Sakamoto et. al. The blue and red residues make up the amino acid binding site, and the red residues are the ones that differ from the wild type ''M. jannaschii'' synthetase that incorporates tyrosine: they determine the specificity for Iodo-tyrosine.[[#References|[3]]] The binding pocket is where mutations are made to incorporate different non-canonical amino acids such as L-DOPA. |
We submitted the tRNA and synthetase pair to incorporate L-DOPA at the UAG codon to the parts registry; the synthetase can be mutated by other teams to incorporate other non-canonical amino acids. | We submitted the tRNA and synthetase pair to incorporate L-DOPA at the UAG codon to the parts registry; the synthetase can be mutated by other teams to incorporate other non-canonical amino acids. | ||
| - | |||
| - | |||
| - | |||
===Efficiency=== | ===Efficiency=== | ||
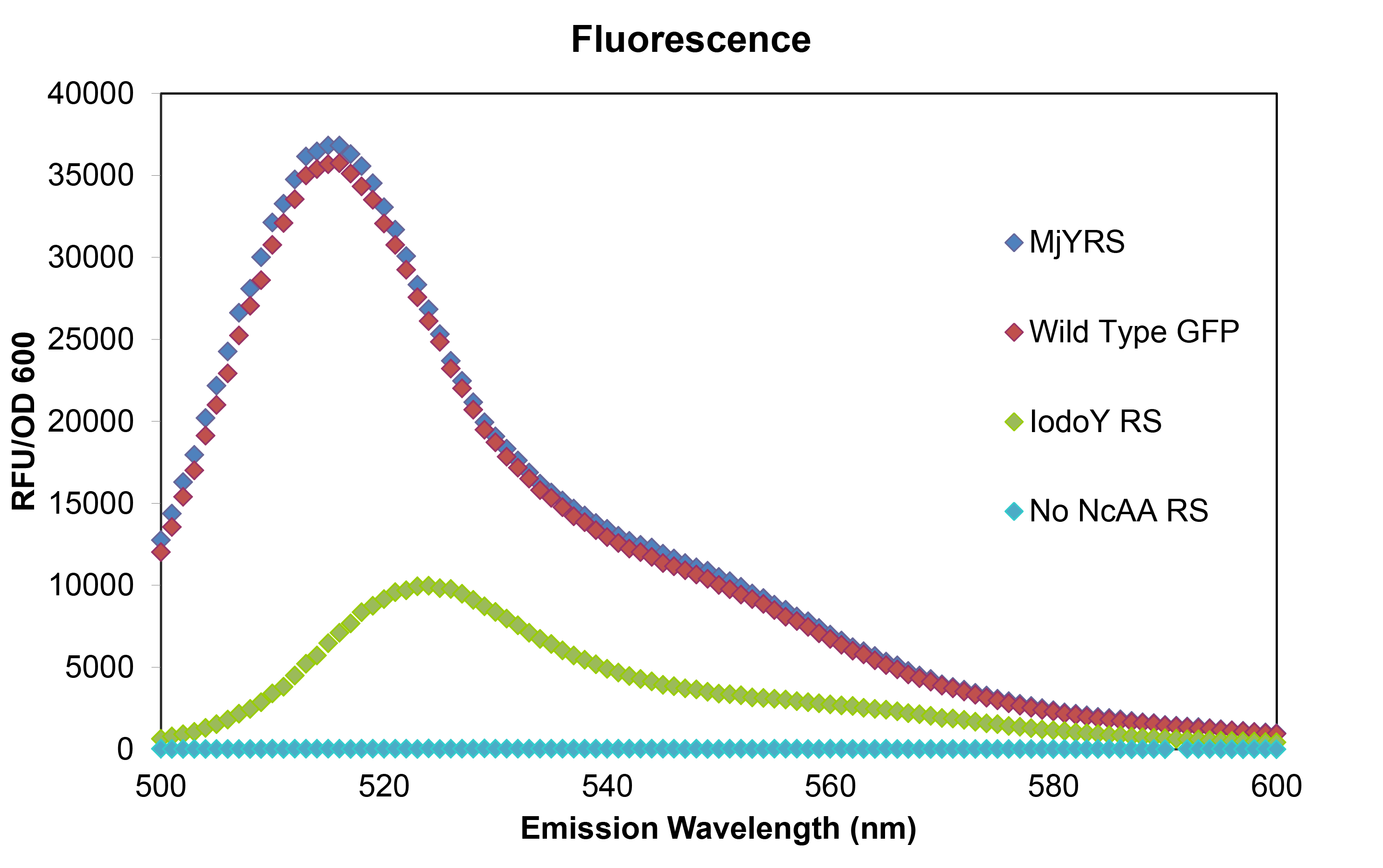
| - | To test that the ''M. jannaschii'' synthetase and tRNA pair actually incorporate a non-canonical amino acid at the Amber stop codon, we designed a | + | To test that the ''M. jannaschii'' synthetase and tRNA pair actually incorporate a non-canonical amino acid at the Amber stop codon, we designed a plasmid containing GFP with an Amber stop codon in place of the tyrosine codon at the active site (Tyr66). If translation stops at this site, then no GFP fluorescence can be seen; if an amino acid other than tyrosine is incorporated at this site, the emission wavelength of the fluorescence may be shifted. We tested this GFP plasmid in ''E. coli'' cells with the wild-type''M. jannaschii'' tyrosyl-tRNA and synthetase, in cells with the ''M. jannaschii'' tRNA and synthetase mutated to incorporate the non-canonical amino acid 3-iodotyrosine at UAG, in cells without an ''M. jannaschii'' synthetase, and compared their fluorescence to the expression of wild type GFP without an Amber stop codon in the active site. |
[[File:Synthetasegfp.png|500px|center]] | [[File:Synthetasegfp.png|500px|center]] | ||
| - | The red curve represents normal wild type GFP fluorescence without an Amber stop codon (Wild Type GFP), the blue curve labeled MjYRS represents fluorescence of the GFP plasmid with the amber stop codon at the active site of GFP in cells with the ''M. jannaschii'' tyrosine synthetase and tRNA (MjYRS), the green curve represents fluorescence of the same GFP plasmid in cells with the ''M. jannaschii'' synthetase and tRNA mutated to incorporate Iodo-tyrosine (IodoY RS), and the blue curve represents fluorescence of the same GFP plasmid in cells without an ''M. jannaschii'' synthetase and tRNA pair (No | + | The red curve represents normal wild type GFP fluorescence without an Amber stop codon (Wild Type GFP), the blue curve labeled MjYRS represents fluorescence of the GFP plasmid with the amber stop codon at the active site of GFP in cells with the ''M. jannaschii'' tyrosine synthetase and tRNA (MjYRS), the green curve represents fluorescence of the same GFP plasmid in cells with the ''M. jannaschii'' synthetase and tRNA mutated to incorporate Iodo-tyrosine (IodoY RS), and the blue curve represents fluorescence of the same GFP plasmid in cells without an ''M. jannaschii'' synthetase and tRNA pair (No ncAA RS). The mutated GFP with tyrosine incorporated at the UAG in the active site has the same intensity and wavelength of fluorescence as wild typre GFP which makes sense since their is no change in the amino acid sequence. The mutated GFP with 3-iodotyrosine incorporated at the active site has lower intensity showing that the ''M. jannaschii'' synthetase is not as efficient at incorporating Iodo-tyrosine, and the 10 nm shift in emission wavelength shows that 3-iodotyrosine is indeed being incorporated into and altering the fluorophore of GFP. All of the variants were carried out in RF0 ''E. coli'' cells except for the variant without an ''M. jannaschii'' synthetase which was done in normal BL21 ''E. coli'' because RF0 cells cannot survive without a tRNA and synthetase pair to code for an amino acid at the UAG Amber stop codon. |
== Mussel adhesion proteins == | == Mussel adhesion proteins == | ||
Revision as of 19:06, 27 September 2013
Contents |
Non-canonical Amino Acids
The "genetic code" refers to the rules by which RNA is translated into the proteins which perform most cellular functions. The genetic code is highly conserved throughout all organisms on earth, with 20 amino acids considered to be "canonical". However, many organisms encode non-canonical amino acids, or will modify residues after translation. Therefore, it is possible to transfer these exceptions to the code into organisms commonly used in molecular biology such as E. coli. This increases the toolbox of reactions and functions available when engineering new proteins. One such recoding method is repurposing one of the stop codons in E. coli. There are three stop codons available, and in E. coli the Amber stop codon, UAG, is used relatively rarely, and therefore presents an excellent target for recoding. In fact, this recoding already occurs in the Archaea Methanocaldococcus jannaschii. M. jannashii has a tyrosyl-tRNA synthetase pair where the tRNA is charged with tyrosine and the anticodon loop recognizes the Amber stop codon. The active site of the orthogonal synthetase can be mutated to charge the tRNA with a non-canonical amino acid instead of tyrosine. [1]
Attempting to recode the Amber codon leads to several complications. There are several essential genes which terminate in the Amber stop codon, and which do not function properly if a non-canonical amino acid is used instead of translation stopping. Additionally, the release factor RF1 is responsible for terminating translation when the ribosome reaches an Amber stop codon, and will compete with the tRNA carrying the non-canonical amino acid. In order to fully recode the Amber codon, the strain RF0 [2] contains an F plasmid with all the essential genes normally ending in the amber codon mutated to end in another stop codon, and has RF1 knocked out to facilitate ncAA incorporation.
The image on the left is the M. jannaschii synthetase mutated to incorporate 3-iodotyrosine by Sakamoto et. al. The blue and red residues make up the amino acid binding site, and the red residues are the ones that differ from the wild type M. jannaschii synthetase that incorporates tyrosine: they determine the specificity for Iodo-tyrosine.[3] The binding pocket is where mutations are made to incorporate different non-canonical amino acids such as L-DOPA.
We submitted the tRNA and synthetase pair to incorporate L-DOPA at the UAG codon to the parts registry; the synthetase can be mutated by other teams to incorporate other non-canonical amino acids.
Efficiency
To test that the M. jannaschii synthetase and tRNA pair actually incorporate a non-canonical amino acid at the Amber stop codon, we designed a plasmid containing GFP with an Amber stop codon in place of the tyrosine codon at the active site (Tyr66). If translation stops at this site, then no GFP fluorescence can be seen; if an amino acid other than tyrosine is incorporated at this site, the emission wavelength of the fluorescence may be shifted. We tested this GFP plasmid in E. coli cells with the wild-typeM. jannaschii tyrosyl-tRNA and synthetase, in cells with the M. jannaschii tRNA and synthetase mutated to incorporate the non-canonical amino acid 3-iodotyrosine at UAG, in cells without an M. jannaschii synthetase, and compared their fluorescence to the expression of wild type GFP without an Amber stop codon in the active site.
The red curve represents normal wild type GFP fluorescence without an Amber stop codon (Wild Type GFP), the blue curve labeled MjYRS represents fluorescence of the GFP plasmid with the amber stop codon at the active site of GFP in cells with the M. jannaschii tyrosine synthetase and tRNA (MjYRS), the green curve represents fluorescence of the same GFP plasmid in cells with the M. jannaschii synthetase and tRNA mutated to incorporate Iodo-tyrosine (IodoY RS), and the blue curve represents fluorescence of the same GFP plasmid in cells without an M. jannaschii synthetase and tRNA pair (No ncAA RS). The mutated GFP with tyrosine incorporated at the UAG in the active site has the same intensity and wavelength of fluorescence as wild typre GFP which makes sense since their is no change in the amino acid sequence. The mutated GFP with 3-iodotyrosine incorporated at the active site has lower intensity showing that the M. jannaschii synthetase is not as efficient at incorporating Iodo-tyrosine, and the 10 nm shift in emission wavelength shows that 3-iodotyrosine is indeed being incorporated into and altering the fluorophore of GFP. All of the variants were carried out in RF0 E. coli cells except for the variant without an M. jannaschii synthetase which was done in normal BL21 E. coli because RF0 cells cannot survive without a tRNA and synthetase pair to code for an amino acid at the UAG Amber stop codon.
Mussel adhesion proteins
The mussel is a marine mollusk that lives in the inter-tidal zone on wave washed rocks. To stand the tidal forces, they must bind themselves to these rocks with their endogenous glue. Mussels have an organ called the foot, which is used to both pull the mussel through terrain and to excrete a structure called the byssus. These byssal threads are made up of collagen and other proteins and are what allow the mussel to attach to surfaces.
The properties of mussel glue make it an attractive target for synthetic improvement; mussel glue allows attachment to many different types of surfaces, both organic and inorganic. It is strong and durable as it is able to withstand the harsh ocean environment. It is made of protein, so it can be easily degraded and is environmentally friendly. Most importantly, however, it is biocompatible (non-immunogenic) and—unlike most glues—it works in aqueous environments.
There are several different proteins that make up the byssal threads, but the ones we’re most interested in are the ones that lie at the interface with the surface to which the mussel adheres. These proteins are responsible for the binding to the substrates and therefore have the most potential for use as a glue.
Adhesion relies on post-translational modifications
All of the different mussel foot proteins have a percentage of their tyrosines post-translationally modified to a molecule called L-DOPA. It turns out that the proteins closer to the adhesion surface have a much higher L-DOPA content, and in fact the L-DOPA content is linearly correlated with adhesiveness.
How MAPs adhere to surfaces
To the right is the sequence of a recombinant mussel adhesion protein, fp-151, that has good adhesive properties. Notably, it has high tyrosine content, denoted by the letter Y.
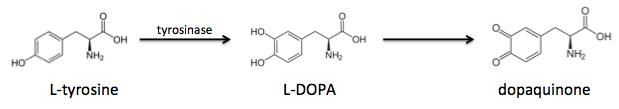
It turns out that tyrosines are critical for adhesion, but first they must undergo tyrosinase-mediated hydroxylation to L-DOPA, which can be further oxidized to dopaquinone:
These modified amino acids contribute to adhesion via two separate mechanisms: L-DOPA interacts non-covalently with inorganic surfaces by chelating metal ions, whereas dopaquinone interacts with organic surfaces through the formation of covalent bonds.
Previous work
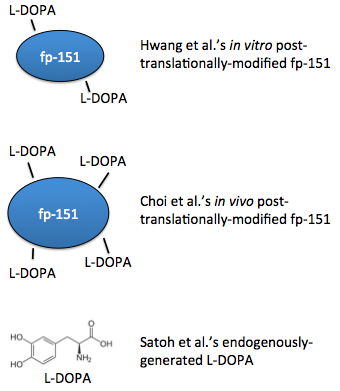
In 2007, a recombinant protein called fp-151 was first produced, which was notable for its lower cytotoxicity than previous recombinant foot proteins. After purifying the protein, tyrosine residues were hydroxylated in vitro with tyrosinase, so the protein was already folded before this modification could occur, possibly limiting access to the tyrosines [4]. Choi, et al. extended this work with a coexpression system, such that the residues were modified in vivo, simultaneously along with translation. These foot proteins had better adhesive properties [5]. Also in 2012, a system to generate L-DOPA from free tyrosine was engineered [6].
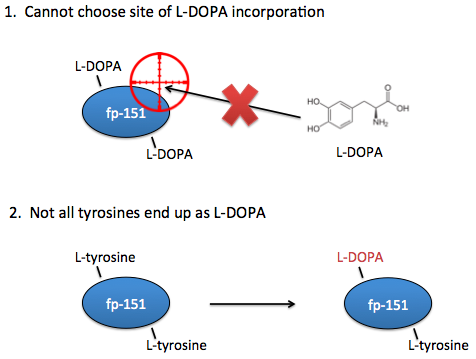
However, these systems are not yet optimal for studying the effects of L-DOPA content on adhesive properties. Ideally, you need the ability to choose exactly which sites that L-DOPA should be incorporated. In current systems, you can only choose where there should be tyrosines.
Additionally, in current systems there is no guarantee that a tyrosine placed at your desired location will even end up as an L-DOPA, so we would also want the ability to know for certain that the residues of interest are in fact L-DOPA and not tyrosine.
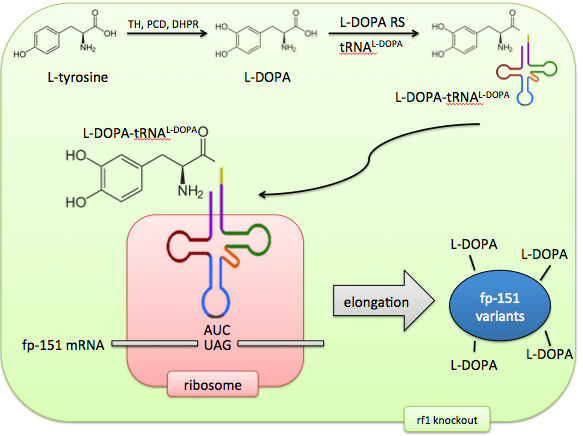
Our solution: direct non-canonical amino acid incorporation
Incorporating the non-canonical amino acid L-DOPA directly would solve these problems of control and predictability. The M. jannaschiitRNA synthetase has been evolved to recognize the non-canonical amino acid L-DOPA by sequential rounds of positive and negative selection. The tRNA’s anticodon loop recognizes the UAG AMber stop codon and therefore this codon is now repurposed to code for a 21st amino acid.
In addition, release factor 1, which normally recognizes this Amber codon, can be knocked out, ensuring no competition between the stop codon’s old effect (termination) and its new effect (L-DOPA incorporation). The result of this system is an expansion of the cell’s genetic code to 21 amino acids, allowing precise control of targeted L-DOPA incorporation.
E. coli as a self-sufficient "black box"
Our vision is of E. coli as a biosynthetic "factory" able to make complicated products with simple inputs. By giving it the machinery to charge tRNA with L-DOPA, and expanding the genetic code to include L-DOPA, this non-canonical amino acid can be incorporated at rationally targetted locations, generating a library of foot protein variants.
Appropriate with this characterization of self-sufficiency and synthetic capability, these E. coli would not even need to be supplemented with exogenous L-DOPA, as they could continually generate it from intracellular free tyrosine.
Experimental design
A two-plasmid system would be appropriate for our aims. One plasmid would code for a library of fp-151 variants, each containing Amber codons in unique locations and quantities, yielding a diverse set of foot proteins with characteristic locations and numbers of L-DOPA residues. The adhesive properties of these variants would be compared against both in vitro and in vivo tyrosinase-treated foot proteins, which would serve as positive controls.
This plasmid would also contain our L-DOPA biosynthesis cassette, ensuring a continual source of non-canonical amino acid without supplementation.
The second plasmid would code for the synthetase/tRNA pair that is responsible for adding L-DOPA to the genetic vocabulary.
A side note: diversity of parts
The individual components of this system are derived from a variety of organisms. The foot proteins come from mussels, the genes in the L-DOPA biosynthesis cassette come from mice and humans, and the synthetase/tRNA pair is derived from archaea, and all these components are put in bacteria.
This covers 5 different organisms from all three domains of life, and as such would be a remarkable illustration of synthetic biology!
Results
Future Directions
References
- Deiters A, and Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg Med Chem Lett. 2005 Mar 1;15(5):1521-4.Pubmed PMID: 15713420.
- Mukai T et. al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010 December; 38(22): 8188–8195. Pubmed PMID: 20702426
- Sakamoto et. al. Genetic Encoding of 3-Iodo-L-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography.Structure. 2009 Mar 11;17(3):335-44. doi: 10.1016/j.str.2009.01.008. PubMed PMID: 19278648.
- Hwang DS, Gim Y, Yoo HJ, Cha HJ. Practical recombinant hybrid mussel bioadhesive fp-151. Biomaterials. 2007 Aug;28(24):3560-8. Epub 2007 May 3. PubMed PMID: 17507090.
- Choi YS, Yang YJ, Yang B, Cha HJ. In vivo modification of tyrosine residues in recombinant mussel adhesive protein by tyrosinase co-expression in Escherichia coli. Microb Cell Fact. 2012 Oct 24;11:139. doi: 10.1186/1475-2859-11-139. PubMed PMID: 23095646; PubMed Central PMCID: PMC3533756.
- Satoh Y, Tajima K, Munekata M, Keasling JD, Lee TS. Engineering of L-tyrosine oxidation in Escherichia coli and microbial production of hydroxytyrosol. Metab Eng. 2012 Nov;14(6):603-10. doi: 10.1016/j.ymben.2012.08.002. Epub 2012 Aug 29. PubMed PMID: 22948011.
 "
"