Team:Greensboro-Austin/Standard Proposal
From 2013.igem.org
Introduction
Previous Standards: As the fields of Synthetic Biology and genetic engineering emerged, arguments for the standardizing the assembly methods involved in constructing plasmids also emerged...
Why standards are awesome... What's great about standards, if followed, is that they allow everyone to be able to construct plasmids and oligos using the same methodology. Once these constructs are submitted into a database it is very easy for others to incorporate those parts into their own constructs because of the standardized submission and assembly methods; thus making collaboration much simpler.
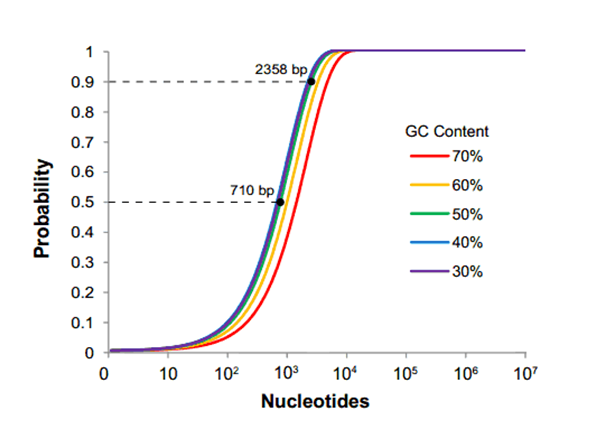
Drawbacks... The issue here is that it requires everyone who wants to submit a part to the database is required to use the same assembly method. This would be totally fine if the standardized assembly method was without fault, but in the case of the Biobrick standard, this simply is not true. It is statistically shown that as the size of your construct approaches around 2400 base pairs, the chance of one of the four restriction sites popping up in your sequence is 90- 100% (as demonstrated by the graph below)
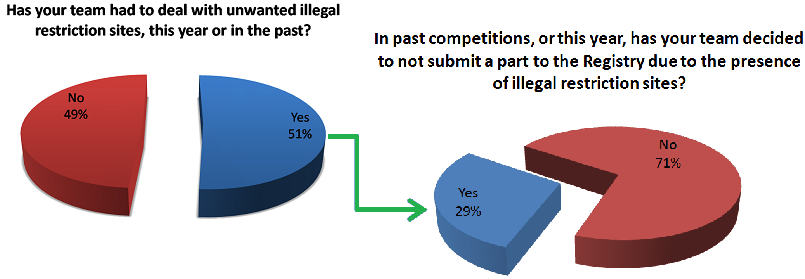
Fixing this problem requires both time and money because in order for anyone to utilize your part using the same standard you need to mutate away those unwanted restriction sites that popped up in your construct. For IGEM teams who have both limited time and funding, this is a real problem and nonetheless an obstacle for any research group trying to share their work using the Biobrick database. Due to these major drawbacks, we speculated that other teams might be having the same issues with regards to part submission and illegal restriction sites. To find out we made a survey asking other teams if they ever come across the same sorts of problems as we had been having and below are some of the results.
Survey Results
As you can see a significant number of our respondents are experiencing the problem of unwanted restriction sites occurring in their parts and thus affecting the number of new parts that are entered into the registry every year. Some notable responses are also shown below.
"I strongly agree with this statement. Having to perform site-directed mutagenesis in order to submit parts to the Registry is a complete waste of time"
"We need to train our students on the way science is being done now, not back in 1980."
"We started our experiments using restriction-ligation but all of these failed. Next we tried Gibson assembly and were almost immediately successful"
"Taking into account the dropping costs of chemical synthesis of DNA, molecular cloning will not be used for biotechnological purposes in a couple of years. Biobrick-based molecular cloning is obsolete."
The Open Sequence Initiative
After many meetings and brainstorming sessions we decided to propose a standard via an RFC that is intended to replace the current Biobrick standard RFC10 in order to reduce restrictions on parts submitted to the Parts Registry. Crucially, this RFC removes the sequence restrictions imposed by RFC10 regarding the removal of the restriction enzymes sites EcoRI, XbaI, SpeI, and PstI from parts before they are eligible for inclusion in the Parts Registry.
Quality Control and Submission Standard Format
Quality control is critical for any serious engineering endeavor. While teams SHOULD sequence verify their parts to detect point mutations and small indels, the BioBricks Foundation needs a simple and rapid method for determining that parts are at least the correct size when submitted to The Registry. Currently this quality control is achieved through the use of the restriction sites present in the BioBrick prefix and suffix. While using these sites for quality control would technically work with this RFC, internal cut sites would result in complex bands when the digest is ran on a gel. Therefore, we propose using the homing endonucleases I-SceI and I-CeuI and their corresponding restriction enzyme sites for quality control purposes. These sites MUST NOT be contained within the part itself, though the probability of this is negligible for gene-sized or even operon-sized parts (Fig. 4).
 "
"