Team:Groningen/Modeling
From 2013.igem.org
| Line 17: | Line 17: | ||
<div align="center"> | <div align="center"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <img src="https://static.igem.org/mediawiki/2013/7/74/AminoAcid_Sequence_Brooks.jpeg" width="80%"> |
</div> | </div> | ||
<p>Figure 1. Amino-acid sequence 2E from A.E.Brooks et al. [5].</p> | <p>Figure 1. Amino-acid sequence 2E from A.E.Brooks et al. [5].</p> | ||
| Line 41: | Line 41: | ||
<div align="center"> | <div align="center"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/8/8b/CodeCogsEqn.gif" width=" | + | <img src="https://static.igem.org/mediawiki/2013/8/8b/CodeCogsEqn.gif" width="20%"> |
</div> | </div> | ||
<p>E.1</p> | <p>E.1</p> | ||
| Line 65: | Line 65: | ||
<div align="center"> | <div align="center"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/f/ff/Error_plot_E2_WithStructure.jpg" width=" | + | <img src="https://static.igem.org/mediawiki/2013/f/ff/Error_plot_E2_WithStructure.jpg" width="160%"> |
</div> | </div> | ||
<p>E.1</p> | <p>E.1</p> | ||
| Line 73: | Line 73: | ||
<div align="center"> | <div align="center"> | ||
| - | <img src="https://static.igem.org/mediawiki/2013/3/34/Error_plot_E2_NoStructure3.jpg" width=" | + | <img src="https://static.igem.org/mediawiki/2013/3/34/Error_plot_E2_NoStructure3.jpg" width="160%"> |
</div> | </div> | ||
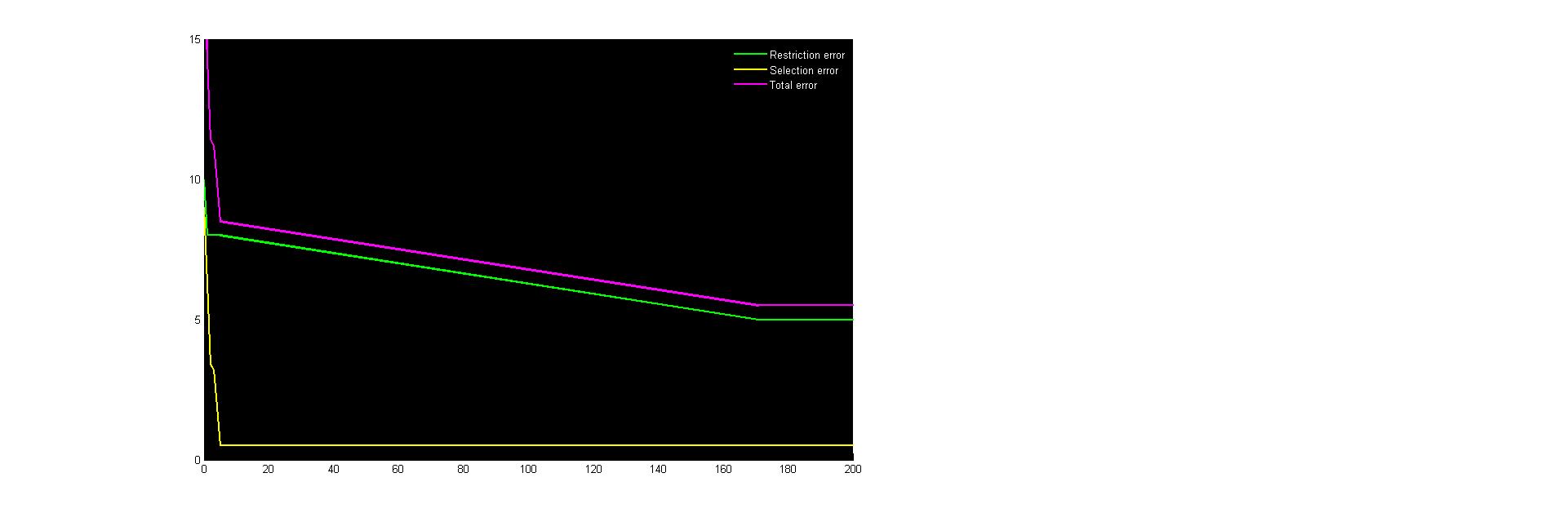
<p>Figure 4. Error plot for sequence 2.</p> | <p>Figure 4. Error plot for sequence 2.</p> | ||
Revision as of 12:36, 24 July 2013
Codon Optimization
The genetic code is degenerate due to the fact that multiple tRNA molecules can be used to encode a single amino-acid. Each protein can therefore be translated from many different nucleotide sequences, which provides us with a degree of control when constructing synthetic genes. The process of finding the optimal sequence for a gene is called codon optimization.
Which sequence is optimal is dependent on the goal, and although codon optimization is mostly used to increase protein expression, it can also be used for inserting or removing certain nucleotide sub-sequences (which are required for functions such as post-translational modifications and DNA degradation), protein folding, and for the modification of protein domains, GC-content, and higher RNA structures.
Goals
The primary goal for applying codon optimization in our project is to increase the expression of the silk gene for maximal silk yield. This can be achieved by increasing both translation elongation as translation initiation rates. The former, the translation elongation rate, is more or less dependent on codon availability, which can form a major bottleneck in the translation process. This bottleneck can be removed, or at least pushed back, by selecting the codons for each amino acid according to some optimal ratio with respect to their availabilities. Besides directly increasing the amino-acid elongation rate, it results in higher tRNA and ribosome recycle rates [1], and has been found to decrease translational errors [2].
Increasing translation initiation rates can be done by increasing mRNA availability, and by taking mRNA secondary structures into account. The former, mRNA availability, can be modified by maximizing mRNA longevity through the removal of endonuclease binding sites. Such sites are recognized by endonuclease proteins, which can then initiate the degradation process of nucleotide sequences. The latter, mRNA secondary structures, can significantly impact protein expression by blocking the ribosome binding cite [3]. There are various mRNA secondary structures, but to keep things simple, we strived for a minimal number of secondary mRNA structures in close proximity to the ribosome binding cite.
Our secondary goal for codon optimization is to help with the secretion of silk. It is known that secretion is, in part, dependent on the correct folding of a protein, and although the folding process is dependent on many different factors, we can exert a degree of control by manipulating translation elongation rates [4]. The goal, then, is to match target and host translational elongation speeds through appropriate codon selection, so as to to reconstruct the folding conditions in the host as closely as possible.
Expression
A 100 amino-acid sequence in bacillus subtilus 168 can potentially have 5^100 unique codon sequences, requiring a calculation time of quindecillions of years! An exhaustive blind search is therefore infeasible, which is why we chose to implement a genetic algorithm in c++.
We initiated the search with a population of 100 semi-random codon sequences (the amino-sequence itself was preserved). Each sequence was then evaluated and given a score based on three factors: (2.1) codon availability, (2.2) RNA secondary structures, and (2.3) endonuclease restriction cites. The top 10% of the population was selected, and was used to rebuild the population with a 2% mutation rate. This cycle of selection and mutation repeated itself for 500 generations. Finally, Due to long calculation times and for convenient future use, we applied the algorithm to a single repeat unit from A.E.Brooks et al. [5] (see figure 1).

Figure 1. Amino-acid sequence 2E from A.E.Brooks et al. [5].
Codon availability
Codon availability, or tRNA availability to be more exact, has been found to be directly proportional to the number of tRNA gene counts - for bacillus a 0.86 correlation was observed [6]. The tRNA gene count data can be found on the online genomic database [7, 8] along with the tRNA usage data, which is the percentage of nucleotide triplets of each codon observed in coding regions. Although the tRNA gene counts give a good impression of tRNA availability, a more accurate representation can be created by incorporating the tRNA usage data.
When adjusting the tRNA availabilities to account for tRNA usage we looked at the standard Watson-Crick and wobble base pairing rules. The first two nucleotide bases bind to anti-codon bases according to the standard Watson-Crick rules; guanine binds with cytosine, and adenine binds with uradine. The third nucleotide, however, can undergo 'wobble', which basically states that the binding rules are less strict [9]. In particular, we adopted the following two rules; tRNA uradine can bind to all mRNA bases, and tRNA guanine can bind to mRNA adenine and uradine.
A final binding rule was adopted due to nucleotide modifications, of which hundreds of different types have been observed. Luckily for us, however, the vast majority of these (over 70%) are adenine to inosine modifications [10]. Moreover, when adenine appears at the wobble position it is almost always converted to inosine [11], which may then bind to mRNA adenine, uradine, and cytosine. The resulting wobble base pairs can be seen in figure 2, where green represents Watson-Crick pairing, blue the permitted wobble pairs, and red the base pairs that are not allowed.

Figure 2
The term 'tRNA usage of codon x' is now perhaps slightly misleading and requires some clarification: since multiple anti-codons can now bind to some codon x, the usage of codon x has to be devided amongst all possible anti-codon matches. To further complicate matters, not all nucleotide bases have the same binding affinity. In particular, we adopted the following two heuristics with respect to wobble base binding preferences; Watson-Crick standard pairs have a higher binding affinity than all wobble base pairs [12], and Inosine-adenine pairs have a lower binding affinity than both inosine-cytosine as inosine-uradine pairs [13].
The formula below shows how the relative codon ratios obtained from the gene count data were adapted. Ai is the availability of codon i, Ux is the usage of codon x, Mx is the number of anti-codon pairs for codon x, and Bi reflects the binding affinity for anti-codon match i.

E.1
The codon availability score for each sequence was then assessed by counting the frequency of each codon, and comparing this to the optimal frequencies obtained from E.1.
RNA secondary structures
Single stranded RNA can fold itself up in many different structures, such as 'hair-pins', 'stem-loops', and 'pseudo-knots'. Such structures can have a significant impact on protein expression, in particular when they occur in close proximity to the ribosome binding cite [3]. When maximizing protein expression, it is therefore advantageous to strive for a minimal amount of RNA secondary structures surrounding the ribosome binding cite.
Predicting these structures, however, is a time-intensive task. We therefore piped the required in and output of our algorithm to, and from, the free-to-use software called RNAStructure [14, 15]. The actual 'RNA secondary structure score' for each sequence was reflected by the number of nucleotide pairs within a range of 90 bases from the ribosome binding cite.
Endonuclease recognition cites
Endonuclease proteins, also known as restriction enzymes, are proteins that are capable of cleaving DNA at specific nucleotide recognition cites. In order to avoid unnecessary cleavage and to increase DNA longevity, these should be removed from the sequence. For bacillus subtilus 168, two recognition cites are currently known: 'GGCC' and 'CTCGAG' [16]. The 'endonuclease score' for each sequence was reflected simply by the number of recognition cite occurrences.
Results
Although the algorithm strives to optimize all three factors mentioned above, at some point one property has to make way for another. Removing a restriction site, for example, may negatively impact the codon selection error, and vica versa. We therefore added different weights to each error value, in order to create two sequences with the following priorities:
Sequence 1: Codon Availability > Secondary RNA Structures > Restriction sites
Sequence 2: Codon Availability > Restriction sites
The idea, then, is to initiate the construct with sequence one, which has a minimal amount of secondary structures at the ribosome binding cite, followed by any number of sequence 2. The error plots of each factor for these sequence 1 and 2 can be seen below. Both were fully optimized at generation ~150, but the calculations continued to generation 500 to make sure.

E.1
Figure 3. Error plot for sequence 1.

Figure 4. Error plot for sequence 2.
C++ code
Our genetic algorithm in C++ coupled with RNAStructure software can be downloaded at "insert link".
Folding
Our secondary goal for codon optimisation is to provide a potential back-up plan for secretion by providing a codon sequence optimized for the correct folding of a protein. It does so by matching translation elongation speeds of the silk proteins MaSp1 and MaSp2 in argiope aurantia and bacillus subtilus 168. Since the translation elongation rate can significantly impact protein folding [4], it may also have an impact on the secretion.
Matching translation elongation rates
In order to match the translation elongation rate of some protein in bacillus subtilus 168 with argiope aurantia, the codon selection should be optimized so as to mirror the corresponding codon availability in argiope aurantia. However, this requires tRNA gene count data of argiope aurantia, which appears to be, as of yet, unavailable. This can be compensated for by the fact that highly expressed genes prefer optimal codons [17], and that MaSp1 and MaSp2 are indeed highly expressed genes.
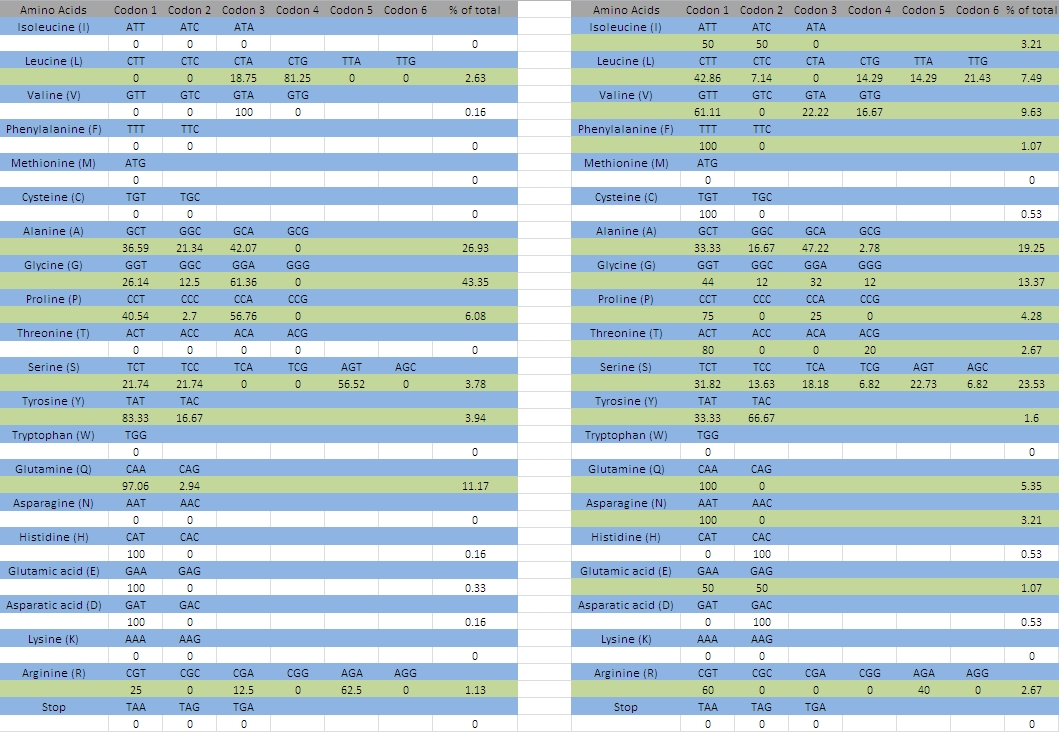
We therefore approximated the optimal selection of tRNA codons by assuming that the main coding regions of MaSp1 and MaSp2 were indeed significantly biased towards optimal codons. The head and tail regions of these genes were excluded from the analysis, since these may be under additional evolutionary pressures, such as the requirement to preserve certain nucleotide sequences for folding. Furthermore, since the total number of codons in MaSp1 and MaSp2 was already relatively small, we excluded all amino-acids constituting less than 1% of the total sequence as these may have a higher chance of being unreliable. The resulting optimal codons selections for each amino-acid can be seen in the left spreadsheet of figure 3, whereas the right spreadsheet shows the same results for the head and the tail. It is clear that the codon bias here is significantly different, which suggests either that there is little to no codon bias in these genes, or that the head and tail regions have indeed undergone additional evolutionary pressures to maintain certain nucleotide sequences.
Figure 5
Blind Search
Because, for protein folding, the correct matching of translation elongation speeds at a local level are more important than correct matching at a global one, we could apply an exhaustive blind search, written in matlab. The optimal codon sequence for each silk protein was the sequence with the smallest codon selection error for each consecutive group of five amino-acids, where the codon selection error was the descrepency in codon availabilites found in bacillus with the availabilities found in argiope aurantia.
References
'''Still have to write the references out'''
[1] Milana Frenkel-Morgenstern, Amar Danon, Thomas Christian, Takao Igarashi, Lydia Cohen, Ya-Ming Hou, Lars Juhl Jensen. (2012). “Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels”. Mol Sys Biol. V8; 572.
[2] http://petrov.stanford.edu/pdfs/65.pdf
[3] Elina Jacobs, James D. Mills, Michael Janitz. 2012. The Role of RNA Structure in
[4] http://www.ncbi.nlm.nih.gov/pubmed/15003273
[5] Brooks
[6] Kanaya S, Yamada Y, Kudo Y, Ikemura T (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143-155.
[7] http://gtrnadb.ucsc.edu/Baci_subt/Baci_subt-summary-codon.html
[8] http://nar.oxfordjournals.org/content/37/suppl_1/D93.long
[9] CRICK, F.H.C.1966.Codon-anticodon pairing: the wobble hypothesis, J.Mol.Biol.19:548-555.
[10] http://books.google.nl/books?id=7BrnbSv4mX8C&pg=PA129&lpg=PA129&dq=RNA+inosine+content&source=bl&ots=M7xihPD9VM&sig=CnQOCIfWA1h5IiIGq1BHSmu-Mtk&hl=nl&sa=X&ei=0Mi4Uc6mJMPtPPTFgAg&ved=0CEkQ6AEwAjgK#v=onepage&q=RNA%20inosine%20content&f=false
[11] http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_2.html
[12] http://www.biomedcentral.com/1471-2148/8/211
[13] http://symposium.cshlp.org/content/47/1087.extract
[14] D.H. Mathews, M.D. Disney, J. L. Childs, S.J. Schroeder, M. Zuker, D.H. Turner (2004). "Incorporating chemical modification constraints into a dynamic programming algorothm for prediction of RNA secondary structure.". Proceedings of the National Academy of Sciences, USA 101 (19): 7287–7292.
[15 D.H. Mathews (2004). "Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization.". RNA 10 (8): 1178–1190.
[16] http://rebase.neb.com/cgi-bin/onumget?845
[17] http://www.ncbi.nlm.nih.gov/pmc/articles/PMC318092/pdf/nar00130-0125.pdf
 "
"