Team:HUST-China/Project
From 2013.igem.org
HUST Oshyn (Talk | contribs) |
HUST Oshyn (Talk | contribs) |
||
| (28 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div id="showcase-cont" class="pr mc h oflowh" style="overflow: visible;"> | <div id="showcase-cont" class="pr mc h oflowh" style="overflow: visible;"> | ||
<div class="showcase-ones" style="display: none; position: relative; left: 0px; top: 0px;" style="left: 0px;"> | <div class="showcase-ones" style="display: none; position: relative; left: 0px; top: 0px;" style="left: 0px;"> | ||
| - | <img class="showcase-left" src="https://static.igem.org/mediawiki/2013/ | + | <img class="showcase-left" src="https://static.igem.org/mediawiki/2013/b/b8/HUST-nav3-2.png" title="" /> |
| - | <img class="showcase-right" src="https://static.igem.org/mediawiki/2013/ | + | <img class="showcase-right" src="https://static.igem.org/mediawiki/2013/5/56/HUST-nav3-1.png" title="" /> |
</div> | </div> | ||
<div class="showcase-ones" style="display: none; position: relative; left: 0px; top: 0px;" style="left: 0px;"> | <div class="showcase-ones" style="display: none; position: relative; left: 0px; top: 0px;" style="left: 0px;"> | ||
| Line 18: | Line 18: | ||
<img class="showcase-right" src="https://static.igem.org/mediawiki/2013/4/4a/HUST-hustchina.png" title="" /> | <img class="showcase-right" src="https://static.igem.org/mediawiki/2013/4/4a/HUST-hustchina.png" title="" /> | ||
</div> | </div> | ||
| + | |||
<div class="showcase-inner none" id="showcase-inner" style="display: block;"> | <div class="showcase-inner none" id="showcase-inner" style="display: block;"> | ||
| Line 33: | Line 34: | ||
<div class="navbar-inner" | <div class="navbar-inner" | ||
style="border-bottom:4px dashed #f2f2f2;"> | style="border-bottom:4px dashed #f2f2f2;"> | ||
| - | <div class="nav-collapse collapse span9 | + | <div style="float:left;"><a href="https://2013.igem.org" title="2013 IGEM Main page"><img src="https://static.igem.org/mediawiki/2013/4/4e/UNIK_Copenhagen_iGEM_logo.png" style="width:50px;"></a></div> |
| + | <div class="nav-collapse collapse span9"> | ||
<ul class="nav"> | <ul class="nav"> | ||
<li> | <li> | ||
| Line 55: | Line 57: | ||
</ul> | </ul> | ||
</div> | </div> | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 71: | Line 73: | ||
<div class="span9" style="padding-left:20px"> | <div class="span9" style="padding-left:20px"> | ||
<div id="scrollspy"> | <div id="scrollspy"> | ||
| + | <!-------------------------------------------main content start--------------------------------------------------> | ||
<div id="Overview"> | <div id="Overview"> | ||
| - | <h1 class="page-header"><strong>Overview</strong></h1> | + | <h1 class="page-header"><strong>Overview</strong></h1> |
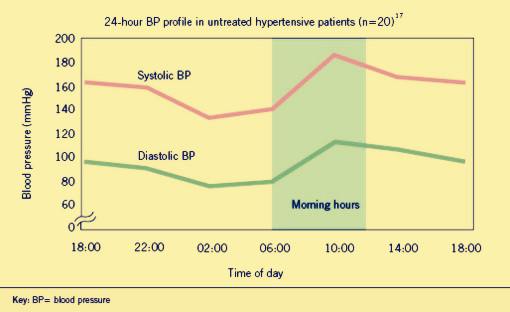
| - | + | <p><b><font size="4px">Hypertension</font></b>is a worldwide public health challenge. And it has been identified as the leading risk factor for mortality mainly because it can lead to adverse cardiovascular events. These events appear to follow a circadian patter, reaching a peak in the morning shortly after wakening and arising. Why? That’s because human’s blood pressure (BP) follows a basic daily rhythm, reaching a peak in the morning when you wake up, then going down and reversing up at 4:00 in the afternoon. (As show in fig.1.1) It is likely that a patient dead in the dream even he felt nothing wrong in the daytime but actually the blood pressure is far beyond the healthy level in the daybreak. Traditional drugs have difficulty to solve the problem as | |
| - | <p> | + | It’s not possible to take pills when asleep. |
| - | + | <br> | |
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
| - | + | <div style="text-align:center"> | |
| - | + | <a href="https://static.igem.org/mediawiki/2013/e/ea/HUST-projectoverview1.jpg" target="_blank"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/e/ea/HUST-projectoverview1.jpg"/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <a href="https://static.igem.org/mediawiki/2013/ | + | |
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | |
</a> | </a> | ||
| - | <p | + | <p><em style="font-size:11px;">Figure1.1: human blood pressure daily rhythm</em></p> |
| - | + | </div> | |
| - | + | <p> | |
| - | + | ||
| - | + | <p>To cope with the situation, our team HUST-China tries to construct a group of friendly probiotic bacteria with the ability of releasing hypotensor in step with human BP daily rhythm in patients’ intestine. In that case, patients will not worry about excessive morning surge anymore. This can be a novel approach to hypertension.</p> | |
| - | + | <p>What can act as the hypotensor? The answer is short chain fatty acid (SCFA). | |
| - | + | SCFA, especially propionate was newly proved to cause an acute hypotensive response by Jennifer et al. A GPCRs called olfr78 expressed in smooth muscle cells of small blood vessel plays an important role. It could be activated by propionate and induce vasodilatation and hypotension. | |
| - | + | </p> | |
| - | + | <p>The question then is: how can we make the hypotensor released periodically?Our solution is bio-oscillator!</p> | |
| - | + | <p>To intervene the bacterium’s generation of propionate. We firstly find a metabolic pathway in E.coil that converts succinate to propionate. (The mechanism of propionate generation is pictured in fig.1.2)There are four enzymes functioning in the pathway. We aimed to find the key gene to be the output of our bio-oscillator.To do this, we recombine the four genes to expression vector, transform them independently and use the four recombinant strains get to do fermentation .Measure propionate concentration by HPLC and find out which gene affects mostly.</p> | |
| - | <a href="https://static.igem.org/mediawiki/2013/ | + | <div style="text-align:center"> |
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <a href="https://static.igem.org/mediawiki/2013/b/b0/HUST-projectoverview3.jpg" target="_blank"> |
| - | + | <img src="https://static.igem.org/mediawiki/2013/b/b0/HUST-projectoverview3.jpg"/> | |
| - | + | </a> | |
| - | + | <p><em style="font-size:11px;">Fig1.2: Mechanism of propionate generation</em></p> | |
| - | + | </div> | |
| - | <a href="https://static.igem.org/mediawiki/2013/ | + | <p>To control the bacterium’s generation of propionate. We use a bio-oscillator. (The design is shown in figure1.3) We firstly use an mRFP as reporter to test and verify our design.</p> |
| - | <img src="https://static.igem.org/mediawiki/2013/ | + | <div style="text-align:center"> |
| - | + | <a href="https://static.igem.org/mediawiki/2013/c/c0/HUST-projectoverview4.png" target="_blank"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/c/c0/HUST-projectoverview4.png"/> | |
| - | + | </a> | |
| + | <p><em style="font-size:11px;"> Figure1.3: Design of our bio-oscillator</em></p> | ||
| + | </div> | ||
| + | <p>After finishing these two main works,we will replace mRFP with our key gene.Regulating the period of oscillator utilizing the frequency divider with an ssrA-tag analog attached to the end of enzyme.<br> | ||
| + | We believe our oscillating BP reliever is both helpful and practical. | ||
| + | </p> | ||
</div> | </div> | ||
<div id="Results"> | <div id="Results"> | ||
<h1 class="page-header"><strong>Results</strong></h1> | <h1 class="page-header"><strong>Results</strong></h1> | ||
| - | + | <p>As we divided our project to two main parts: propionate generation and oscillator. The results will be demonstrated from these two sections.</p> | |
| - | + | <h4>Propionate generation:</h4> | |
| - | < | + | <p>1.We firstly successfully copied the four genes ygfD/ygfH/ygfG/Sbm from E.coli strain K12, and then recombined them independently to expression vector. Purified protein from cell disruptions by Ni-chelating and did gel analysis. From figure-1 , we confirmed that the enzymes were successfully expressed.</p> |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/6/6e/HUST-proj-res1.png" /></p> | |
| - | + | <p><em style="font-size:11px;">Figure1. SDS-PAGE analysis of Sbm, ygfD, ygfG, ygfH over-expression</em></p> | |
| - | + | </div> | |
| - | + | <p>2.As we thought the overexpression of enzymes didn’t directly prove the increase of propionate, and we wanted to find the key gene in the reaction. So we used the positive clones to do fermentation. We firstly optimized the condition and found the best sampling time points. Then we used standard propionate of seven gradients to draw a linear graph between propionate concentration and HPLC peak area. Figure-2 shows the relationship.</p> | |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/7/76/HUST-proj-res2.png" /></p> | |
| - | + | <p><em style="font-size:11px;">Figure-2. Standard curve of propionate and HPLC peak area</em></p> | |
| - | + | </div> | |
| - | + | <p>Later, we used the recombinant strain to do fermentation, and measured propionate concentration in the samples. Figure-3 shows the propionate increase percent of each gene recombinant strain. From the data, we found propionate production had a significant increase when ygfD transformed. In other words, we found the key gene- our oscillator’s output.</p> | |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/0/05/HUST-proj-res3.png" /></p> | |
| - | + | <p><em style="font-size:11px;">Figure-3. HPLC analysis wild-type BL21 and recombination BL21 with four genes</em></p> | |
| - | + | </div> | |
| - | + | <h4>Oscillator:</h4> | |
| - | + | <p>1.As for the oscillator, we first successfully constructed the two plasmids formed the dual-feedback circuit. (Circuit and plasmid were show in figure-4) LAA tag was added to the protein for rapid degradation. Gene sequencing confirmed the plasmids to be correct without any lethal mutation.</p> | |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/0/08/HUST-proj-res4.png" /></p> | |
| - | + | <p>Figure-4.The dual-feedback circuit and two plasmids we constructed</p> | |
| - | + | </div> | |
| - | + | <p>2.After that, we transformed recombinant plasmid pET-28a (+) which had an mRFP reporter to check if it could function well. We induced the positive clone with 2mM IPTG and observed by fluorescence microscope. Figure-5 shows that it function well.</p> | |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/8/83/HUST-proj-res5.png" /></p> | |
| - | + | <p><em style="font-size:11px;">Figure-5. Fluorescence microscope photo of recombinant plasmid pET-28a (+) transformed cell induced by 2mM IPTG</em></p> | |
| - | + | </div> | |
| - | + | <p>3.Later, we co-transformed two recombinant plasmids pET-28a (+) and pACYCDuet-1. After inducing positive clone by 0.7% Arabinose and 2 mM IPTG, we used fluorescence microscope to see RFP change of single cell and used fluorospectro photometer to see RFP change of multicells.Figure-6 & Figure-7 shows the result.</p> | |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/e/ec/HUST-proj-res6.png" /></p> | |
| - | + | <p><em style="font-size:11px;">Figure-6. Fluorescence change of single cell</em></p> | |
| - | + | </div> | |
| - | + | <p>Cells were diluted with culture medium and immobilized with glycerol. (5ul bacteria +20ulmedium+20ul glycerol). Making sure that the cell was alive and motionless, we could take photo of the same cell. From the pictures, we can see that the fluorescence changed with time in an oscillatory way, which supports our bio-oscillator design.</p> | |
| - | + | <div style="text-align:center;"><p><img src="https://static.igem.org/mediawiki/2013/0/01/HUST-proj-res7.jpg" /></p> | |
| - | + | <p><em style="font-size:11px;">Figure-7. Fluorescence change of multicell</em></p> | |
| - | + | </div> | |
| - | + | <p>We used fluorospectro photometer to measure the oscillating behavior of multi-cells. | |
| + | After induced by 0.7% arabinose and 2 mM IPTG when OD600 was 0.55, cells were cultured in 37℃/200rpm.We sampled a series of time points and draw the curve as figure-7. We saw a significant fluorescence oscillating in compare to control group. The control group cells transformed pET-28a (+) only.</p> | ||
| + | <p>Summarization: We have successfully found a way to enhance cells generation of propionate, tested and verified our design of bio-oscillator. Combining these two works, we believe we can build a gut probiotic which can release propionate periodically in accord with the rhythm of human BP.</p> | ||
| + | |||
</div> | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<div id="Future-Work"> | <div id="Future-Work"> | ||
<h1 class="page-header"><strong>Future work</strong></h1> | <h1 class="page-header"><strong>Future work</strong></h1> | ||
| - | <p> | + | <p>We do not have enough time to fulfill the whole project. Based on the pre-existing work, these tasks are coming to be finished in future.<br> |
| - | + | 1.Replace the report gene mRFP by ygfD in the dual-feedback circuit to see whether the propionate generation can oscillate.<br> | |
| - | 1. | + | 2.Regulate the period of propionate generation to cope with human’s BP rhythm. We may utilize the frequency divider with an ssrA-tag analog attached to the end of enzyme to achieve it.<br> |
| - | + | <div style="float:right"><img src="https://static.igem.org/mediawiki/2013/d/d6/HUST-proj-future-work.png" width="200" /></div> | |
| - | 3. | + | 3.Transform the regulatory net into bifidobacterium – microorganism has reputation among the dairy industry — due to people who we sent questionnaire to show preference eating food containing probiotics rather than E.coli, Also, we will measure the propionate outside of the human body.<br> |
| - | + | ||
</p> | </p> | ||
| Line 174: | Line 168: | ||
<h1 class="page-header"><strong>Judging Critieria</strong></h1> | <h1 class="page-header"><strong>Judging Critieria</strong></h1> | ||
<p>Already registered in the official website in 13th March and was accepted in 12th April. <br> | <p>Already registered in the official website in 13th March and was accepted in 12th April. <br> | ||
| - | We completed safety form, judging form and team wiki before the deadline. It is for sure that we are going to present a poster and a talk at the iGEM Jamboree. <br> | + | 1.We completed safety form, judging form and team wiki before the deadline. It is for sure that we are going to present a poster and a talk at the iGEM Jamboree. <br> |
| - | We documented four newly standard BioBrick Part(sbm/ygfG/ygfH/ygfD) used in our project and submitted them to the iGEM Registry adhere to guidelines. <br> | + | 2.We documented four newly standard BioBrick Part(sbm/ygfG/ygfH/ygfD) used in our project and submitted them to the iGEM Registry adhere to guidelines. <br> |
| - | Our works aims at maintaining the blood pressure through microbe metabolism SCFA, which is a new application in medicine to our knowledge.<br> | + | 3.Our works aims at maintaining the blood pressure through microbe metabolism SCFA, which is a new application in medicine to our knowledge.<br> |
| - | We did plenty of experiment to validate that two of BioBrick Part of our own design and construction works as expected. <br> | + | 4.We did plenty of experiment to validate that two of BioBrick Part of our own design and construction works as expected. <br> |
| - | We share information with WHU and HZAU | + | 5.We share information and material with WHU and HZAU .Cooperating with HZAU on characterizing one part.<br> |
| + | 6.We originaly creat a crossword to popularize historical knowledge about iGEM. That's a good new approach for human practice.<br> | ||
| + | Therefore, we believe that we deserve a Gold Medal Prize.<br> | ||
</p> | </p> | ||
| - | </div> | + | </div> |
| + | <!-------------------------------------------main content end----------------------------> | ||
</div> | </div> | ||
| Line 188: | Line 185: | ||
</div> | </div> | ||
| - | <div class="modal-footer footer"> | + | <div class="modal-footer footer"><br><br> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<div> | <div> | ||
CopyRight ©<a href="https://2013.igem.org">2013.igem.org</a> All Rights Reserved! | CopyRight ©<a href="https://2013.igem.org">2013.igem.org</a> All Rights Reserved! | ||
| Line 218: | Line 203: | ||
var item3= $("#Future-Work").outerHeight(); | var item3= $("#Future-Work").outerHeight(); | ||
var item4= $("#Judging-Criteria").outerHeight(); | var item4= $("#Judging-Criteria").outerHeight(); | ||
| - | if(scroll > | + | if(scroll > 204){ |
$("#up").slideDown("ease-out"); | $("#up").slideDown("ease-out"); | ||
$("#mainNavBar").addClass("navbar-fixed-top"); | $("#mainNavBar").addClass("navbar-fixed-top"); | ||
$(".bs-docs-sidenav").removeClass("affix-top").addClass("affix"); | $(".bs-docs-sidenav").removeClass("affix-top").addClass("affix"); | ||
| - | if (scroll < | + | if (scroll < 204 + item1){ |
navli.removeClass("active").eq(0).addClass("active"); | navli.removeClass("active").eq(0).addClass("active"); | ||
| - | }else if (!(scroll < | + | }else if (!(scroll < 204 + item1|| scroll > 204 + item1+ item2)){ |
navli.removeClass("active").eq(1).addClass("active"); | navli.removeClass("active").eq(1).addClass("active"); | ||
| - | }else if (!(scroll < | + | }else if (!(scroll < 204 + item1+ item2|| scroll > 204 + item1+ item2+ item3)){ |
navli.removeClass("active").eq(2).addClass("active"); | navli.removeClass("active").eq(2).addClass("active"); | ||
| - | }else if (!(scroll < | + | }else if (!(scroll < 204 + item1+ item2+ item3|| scroll > 204 + item1+ item2+ item3+ item4)){ |
navli.removeClass("active").eq(3).addClass("active"); | navli.removeClass("active").eq(3).addClass("active"); | ||
} | } | ||
Latest revision as of 13:58, 28 October 2013
Overview
Hypertensionis a worldwide public health challenge. And it has been identified as the leading risk factor for mortality mainly because it can lead to adverse cardiovascular events. These events appear to follow a circadian patter, reaching a peak in the morning shortly after wakening and arising. Why? That’s because human’s blood pressure (BP) follows a basic daily rhythm, reaching a peak in the morning when you wake up, then going down and reversing up at 4:00 in the afternoon. (As show in fig.1.1) It is likely that a patient dead in the dream even he felt nothing wrong in the daytime but actually the blood pressure is far beyond the healthy level in the daybreak. Traditional drugs have difficulty to solve the problem as
It’s not possible to take pills when asleep.
To cope with the situation, our team HUST-China tries to construct a group of friendly probiotic bacteria with the ability of releasing hypotensor in step with human BP daily rhythm in patients’ intestine. In that case, patients will not worry about excessive morning surge anymore. This can be a novel approach to hypertension.
What can act as the hypotensor? The answer is short chain fatty acid (SCFA). SCFA, especially propionate was newly proved to cause an acute hypotensive response by Jennifer et al. A GPCRs called olfr78 expressed in smooth muscle cells of small blood vessel plays an important role. It could be activated by propionate and induce vasodilatation and hypotension.
The question then is: how can we make the hypotensor released periodically?Our solution is bio-oscillator!
To intervene the bacterium’s generation of propionate. We firstly find a metabolic pathway in E.coil that converts succinate to propionate. (The mechanism of propionate generation is pictured in fig.1.2)There are four enzymes functioning in the pathway. We aimed to find the key gene to be the output of our bio-oscillator.To do this, we recombine the four genes to expression vector, transform them independently and use the four recombinant strains get to do fermentation .Measure propionate concentration by HPLC and find out which gene affects mostly.
To control the bacterium’s generation of propionate. We use a bio-oscillator. (The design is shown in figure1.3) We firstly use an mRFP as reporter to test and verify our design.
After finishing these two main works,we will replace mRFP with our key gene.Regulating the period of oscillator utilizing the frequency divider with an ssrA-tag analog attached to the end of enzyme.
We believe our oscillating BP reliever is both helpful and practical.
Results
As we divided our project to two main parts: propionate generation and oscillator. The results will be demonstrated from these two sections.
Propionate generation:
1.We firstly successfully copied the four genes ygfD/ygfH/ygfG/Sbm from E.coli strain K12, and then recombined them independently to expression vector. Purified protein from cell disruptions by Ni-chelating and did gel analysis. From figure-1 , we confirmed that the enzymes were successfully expressed.

Figure1. SDS-PAGE analysis of Sbm, ygfD, ygfG, ygfH over-expression
2.As we thought the overexpression of enzymes didn’t directly prove the increase of propionate, and we wanted to find the key gene in the reaction. So we used the positive clones to do fermentation. We firstly optimized the condition and found the best sampling time points. Then we used standard propionate of seven gradients to draw a linear graph between propionate concentration and HPLC peak area. Figure-2 shows the relationship.

Figure-2. Standard curve of propionate and HPLC peak area
Later, we used the recombinant strain to do fermentation, and measured propionate concentration in the samples. Figure-3 shows the propionate increase percent of each gene recombinant strain. From the data, we found propionate production had a significant increase when ygfD transformed. In other words, we found the key gene- our oscillator’s output.

Figure-3. HPLC analysis wild-type BL21 and recombination BL21 with four genes
Oscillator:
1.As for the oscillator, we first successfully constructed the two plasmids formed the dual-feedback circuit. (Circuit and plasmid were show in figure-4) LAA tag was added to the protein for rapid degradation. Gene sequencing confirmed the plasmids to be correct without any lethal mutation.

Figure-4.The dual-feedback circuit and two plasmids we constructed
2.After that, we transformed recombinant plasmid pET-28a (+) which had an mRFP reporter to check if it could function well. We induced the positive clone with 2mM IPTG and observed by fluorescence microscope. Figure-5 shows that it function well.

Figure-5. Fluorescence microscope photo of recombinant plasmid pET-28a (+) transformed cell induced by 2mM IPTG
3.Later, we co-transformed two recombinant plasmids pET-28a (+) and pACYCDuet-1. After inducing positive clone by 0.7% Arabinose and 2 mM IPTG, we used fluorescence microscope to see RFP change of single cell and used fluorospectro photometer to see RFP change of multicells.Figure-6 & Figure-7 shows the result.

Figure-6. Fluorescence change of single cell
Cells were diluted with culture medium and immobilized with glycerol. (5ul bacteria +20ulmedium+20ul glycerol). Making sure that the cell was alive and motionless, we could take photo of the same cell. From the pictures, we can see that the fluorescence changed with time in an oscillatory way, which supports our bio-oscillator design.

Figure-7. Fluorescence change of multicell
We used fluorospectro photometer to measure the oscillating behavior of multi-cells. After induced by 0.7% arabinose and 2 mM IPTG when OD600 was 0.55, cells were cultured in 37℃/200rpm.We sampled a series of time points and draw the curve as figure-7. We saw a significant fluorescence oscillating in compare to control group. The control group cells transformed pET-28a (+) only.
Summarization: We have successfully found a way to enhance cells generation of propionate, tested and verified our design of bio-oscillator. Combining these two works, we believe we can build a gut probiotic which can release propionate periodically in accord with the rhythm of human BP.
Future work
We do not have enough time to fulfill the whole project. Based on the pre-existing work, these tasks are coming to be finished in future.
1.Replace the report gene mRFP by ygfD in the dual-feedback circuit to see whether the propionate generation can oscillate.
2.Regulate the period of propionate generation to cope with human’s BP rhythm. We may utilize the frequency divider with an ssrA-tag analog attached to the end of enzyme to achieve it.

Judging Critieria
Already registered in the official website in 13th March and was accepted in 12th April.
1.We completed safety form, judging form and team wiki before the deadline. It is for sure that we are going to present a poster and a talk at the iGEM Jamboree.
2.We documented four newly standard BioBrick Part(sbm/ygfG/ygfH/ygfD) used in our project and submitted them to the iGEM Registry adhere to guidelines.
3.Our works aims at maintaining the blood pressure through microbe metabolism SCFA, which is a new application in medicine to our knowledge.
4.We did plenty of experiment to validate that two of BioBrick Part of our own design and construction works as expected.
5.We share information and material with WHU and HZAU .Cooperating with HZAU on characterizing one part.
6.We originaly creat a crossword to popularize historical knowledge about iGEM. That's a good new approach for human practice.
Therefore, we believe that we deserve a Gold Medal Prize.
 "
"