Team:HUST-China/Project
From 2013.igem.org
Overview
Hypertension is an important worldwide public-health challenge.It has been identified as the leading risk factor for mortality mainly because it can lead to adverse cardiovascular events. It has already affected one billion people through out world and killed nine million people each year. And the incidence of it appears to follow a circadian pattern, reaching a peak in the morning shortly after wakening and arising.
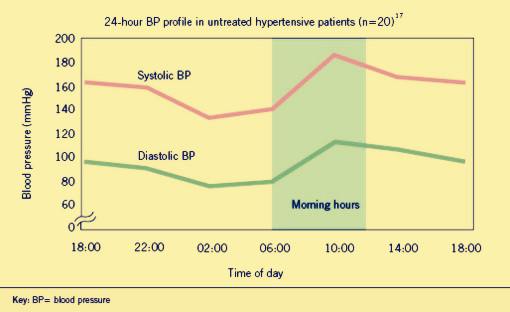
There is a wide selection of antihypertensive drugs designed for patients. However, traditional treatment comes along with heavy financial burden and severe side effects along with drug dependence. To make the matter worse, these medicines can not stop morning surge from taking patients' lives.
Short chain fatty acid(SCFA) ,especially acetate and propionate, was newly proved to cause an acute hypotensive response. A GPCRs called olfr78 expressed in smooth muscle cells of small blood vessel plays an important role. It could be activated by SCFA and induce vasodilatation and hypotension. In consideration of the case, we developed a novel method to treat hypertension by utilizing a group of friendly engineering bacteria which can release propionate periodically in human intestine.
To achieve this,we found a four-enzyme pathway in E.coil that converts succinate to propionate. By combining key genes with bio-oscillator, we try to make E.coli release propionate in patients’intestine periodically. Once the E.coli is delivered into human body as probiotics, the propionate can be taken by the circulatory system and act with the receptors. In that case, patients will not worry about excessive surge anymore.
Results
1 .Biobricks (we submit / Registry ) table describe
| Standard biobricks | description |
|---|---|
| pSB1C3-ygfD(EocRI PstI) | According to Toomas Haller, the enzyme encoded by the second gene- ygfD, contains a consensus binding sequence for ATP. They thought it might be a succinate (or propionate)CoA ligase, or a novel (biotin-independent) propionyl-CoA carboxylase. |
| pSB1C3-ygfH(EocRI PstI) | This gene encode propionyl-CoA:succinate CoA transferase that catalyzes a CoA transferase reaction from propionyl-CoA to succinyl, generating propionate. |
| pSB1C3-ygfG(EocRI PstI) | The third gene in the operon encoding methylmalonyl-CoA decarboxylase that catalyzes the decarboxylation of methylmalonyl-CoA to propionyl-CoA |
| pSB1C3-sbm(EocRI PstI) | Sbm encodes methylmalonyl-CoA epimerase which catalyzes the reversible reaction of succinyl-CoA and methylmalonyl -CoA |

3.As to the oscillator device, the dual-feedback circuit driven by the hybrid promoter was successfully constructed.


Future work
There is a big challenge in our project that the recombination microbe has to be validated in the human intestine. Based on the pre-existing work, a plenty of works are coming to us in the near future.
1.Regulating the period of propionate utilizing the frequency divider with a ssrA-tag analog attached to the end of enzyme.
2. Replace the report gene rfp encoding red fluorescence protein with key gene encoding enzymes in the synthetic pathway.
3. As we know that E.coli is the most popular chassi used in the synthetic biology. But according to the people we sent questionnaire to, they prefer eating food containing probiotics rather than eating bacteria. So we are going to transform the regulatory net into bifidobacterium, which enjoy a highly reputation among the dairy industry. We will measure the propionate outside of the human body.
4. Using mathematical modeling to imitate the environment in the intestine and the concentration decreasing of propionate in the blood circulation.
Judging Critieria
Already registered in the official website in 13th March and was accepted in 12th April.
1.We completed safety form, judging form and team wiki before the deadline. It is for sure that we are going to present a poster and a talk at the iGEM Jamboree.
2.We documented four newly standard BioBrick Part(sbm/ygfG/ygfH/ygfD) used in our project and submitted them to the iGEM Registry adhere to guidelines.
3.Our works aims at maintaining the blood pressure through microbe metabolism SCFA, which is a new application in medicine to our knowledge.
4.We did plenty of experiment to validate that two of BioBrick Part of our own design and construction works as expected.
5.We share information and material with WHU and HZAU .Cooperating with HZAU on characterizing one part.
6.We originaly creat a crossword to popularize historical knowledge about iGEM. That's a good new approach for human practice.
Therefore, we believe that we deserve a Gold Medal Prize.
 "
"











