Team:Heidelberg/Templates/DelH week10
From 2013.igem.org
01-07 - 07-07-13
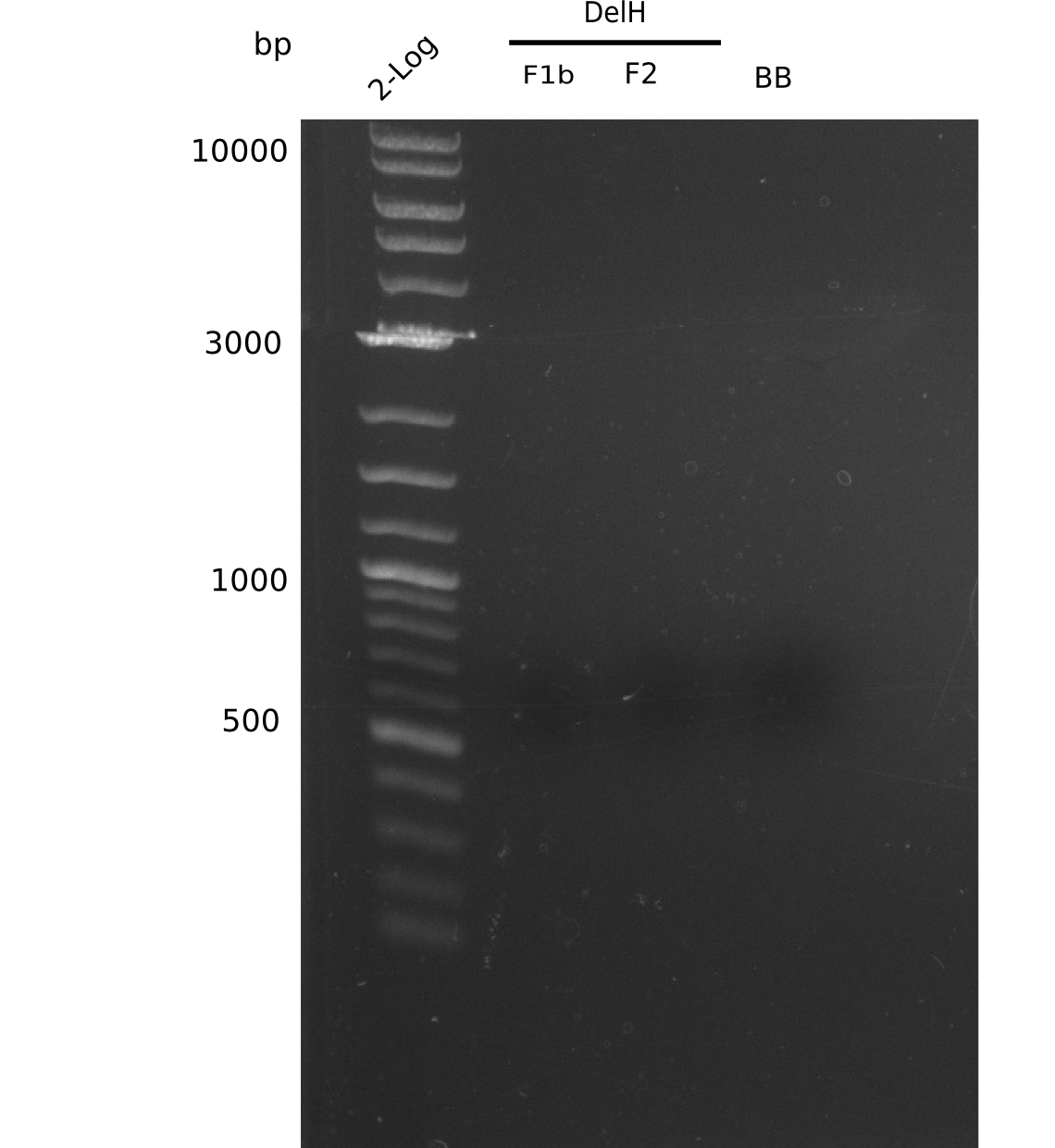
Amplification of DelH F1b
PCR Conditions F1b.W10.A
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl DelH 1Fb Fragment |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 30 |
| 30 | 98 | 15 |
| 68 | 15 | |
| 72 | 5 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
No correct bands visible.
- => Phusion is no option for us, not even for re-PCRs.
PCR Conditions F1b.W10.B
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 180 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 5 Kb
- => F1b was not amplified.
Amplification of DelH F2
PCR Conditions F2.W10.A
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl DelH 2 PCR product |
| Primer fw 10 µM | 2.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f2_KpnI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 15 |
| 66 decrease by 0.5 | 15 | |
| 72 | 8 min | |
| 12 | 98 | 15 |
| 66 | 15 s | |
| 72 | 8 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb, Loaded 1 µl of PCR
Gel does not show correct band.
- => Phusion is no option for us, not even for re-PCRs.
PCR Conditions F2.W10.B
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl D. acidovorans glycerol stock |
| Primer fw 10 µM | 2.5 µl DelH_f2_SalI_fw 10 µM |
| Primer rev 10 µM | 2.5 µl DelH_f2_KpnI_rev 10 µM |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 12 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 8 Kb, Loaded 50 µl of PCR
- => F2 was not amplified.
Amplification of Backbone
PCR Conditions BB.W10.A
| Reagent | Backbone |
|---|---|
| Template | 1 µl psB6A1+AraC+Lacz 1:10 |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 15 | |
| 72 | 8 min | |
| 12 | 98 | 15 |
| 66 | 15 | |
| 72 | 8 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb, Loaded 1 µl of PCR
No product.
- => Phusion is no option for us, not even for re-PCRs.
PCR Conditions BB.W10.B
| Reagent | Backbone |
|---|---|
| Template | 1 µl psB6A1+AraC+Lacz 1:10 |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 12 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
- Using hot start at 98°C
Result
Expected band: 7.3 Kb
- => BB was not amplified completely. It seems that there is no insert present or the conditions were not optimal. Repeat the PCR with other conditions.
PCR Conditions BB.W10.C
| Reagent | Backbone |
|---|---|
| Template | 1 µl psB6A1+AraC+Lacz 1:10 |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles-PCR | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 180 |
| 1 | 4 | inf |
Result
Expected bands: 7.3 Kb, Loaded 50 µl of PCR
Expected band is there, but also unspecific ones.
- => Correct band was cut and gel extracted.
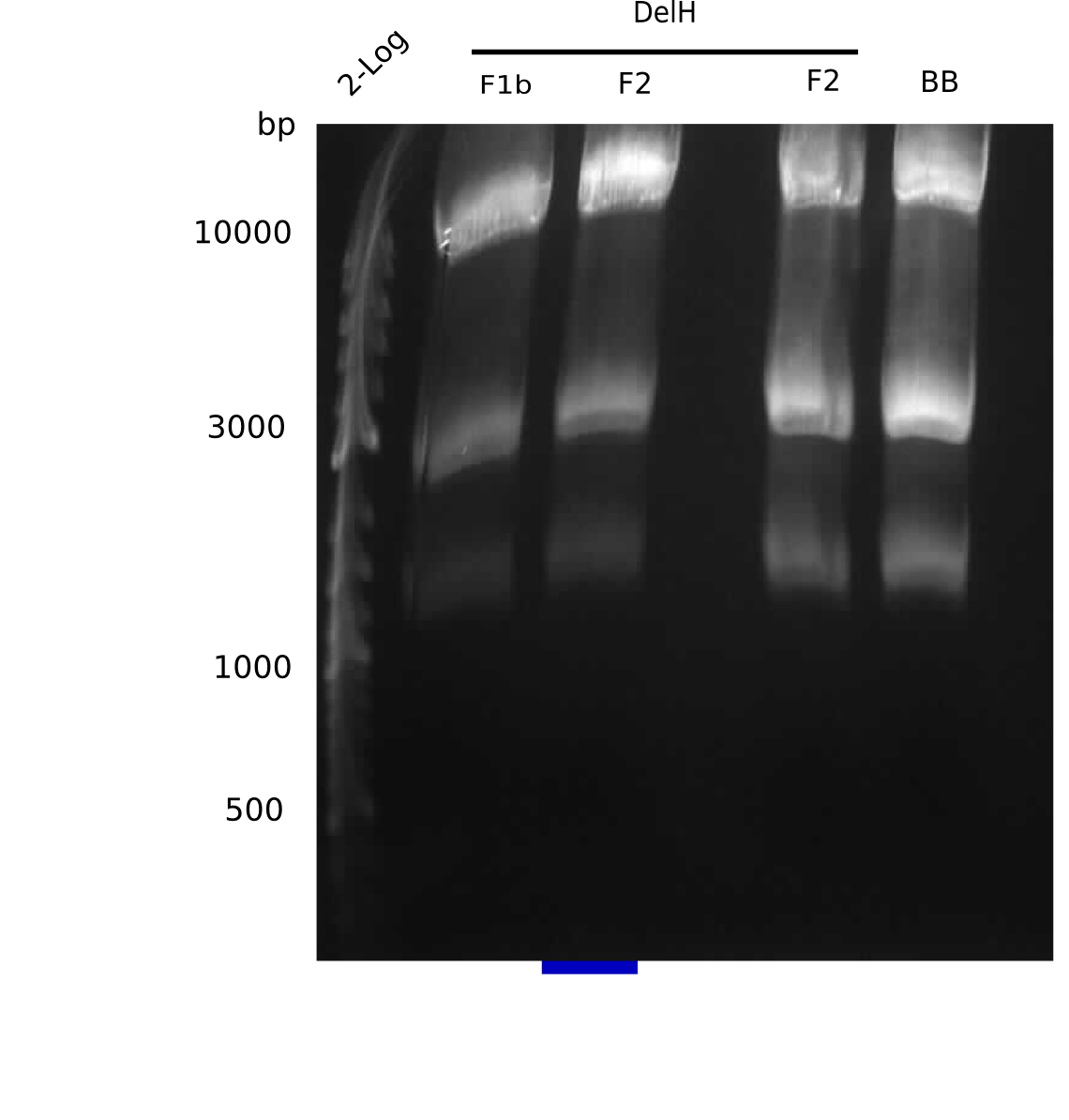
Generation of DelH Plasmid
Current Status
| Fragment | Concentration [ng/µl] | Digested and Purified |
|---|---|---|
| F1a | 267 | check |
| F1b | 9.8 | - |
| F2 | 11 (gel extracted) and 369 (PCR purified) | - |
| pSB6A1-AraC-lacZ | 53.5 and 38.5 | - |
Restriction Digest of Fragments
| Fragment | Amount for Digest [ng] | Enzymes [µl] | Buffer [µl] |
|---|---|---|---|
| F1b | 441 |
EcoRI-HF: 1 | Cut smart 5.5 |
| F2 | 495 |
KpnI: 1 | Buffer 2.1: 5.5 |
| pSB6A1-AraC-lacZ | 1,599 |
KpnI: 1 | Buffer 1.1: 3.5 |
- 1 h at 37°C
Isopropanol Purification
| Fragment | Concentration after Digest & Purification [ng/µl] | Sequences |
|---|---|---|
| F1a | 267 |
File:Heidelberg PCR(DelH) restricted(KpnI+PacI) sequence.fasta.txt |
| F1b | 3.5 | (Entire) Sequence of DelH |
| F2 | 1.8 | |
| pSB6A1-AraC-lacZ | 7.3 |
File:Heidelberg PCR(pSB6A1-AraC-lacZ) digest.fasta.txt |
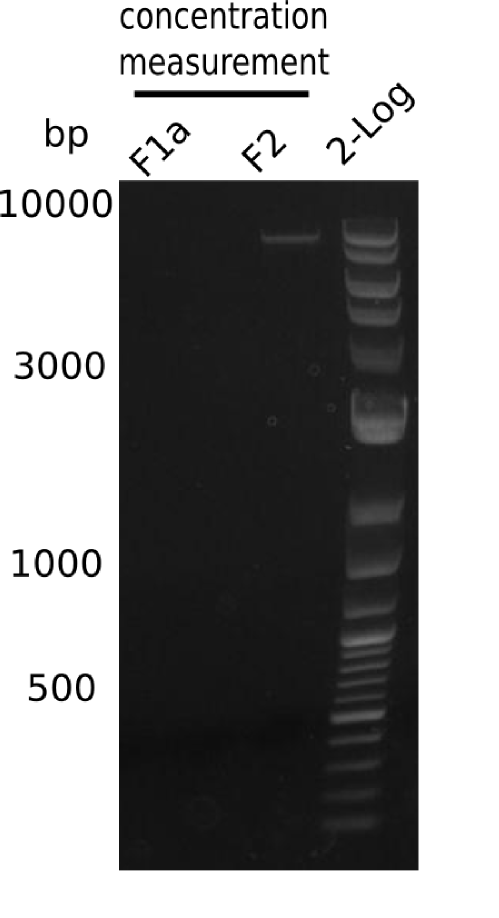
Confirmation of DNA Concentration
| Fragment | Concentration [ng/µl] | Date | µl on gel | Expected band size |
|---|---|---|---|---|
| F1a | 261 | 19-06-13 | 1 µl | 5 Kb |
| F1a | 261 | 19-06-13 | 2 µl | 5 Kb |
| F2 | 369 | 01-07-13 | 1 µl | 8 Kb |
Fragment F1a is not present, all others are fine.
- => Therefore, the next steps are:
- 1. Measurement of concentration = 40 ng/µl
- 2. PCR of the fragment F1a. As template, we use the purified PCR with the concentration of 40 ng/µl.
Restriction Digest of F2 and Backbone
| Fragment | Volume for digest [ng] | Enzymes [µl] | Buffer [µl] |
|---|---|---|---|
| 2 | 40*369 = 14.76 µg |
KpnI: 1 | Buffer 2.1: 4.8 |
| pSB6A1-AraC-lacZ | 40*38.5 = 1.54 µg |
KpnI: 1 | Buffer 1.1: 1.8 |
- 1h at 37°C
Purification of Restriction Digest
Using nucleotide removal kit (Qiagen), eluted with 30 µl ddH2O.
| Fragment | Concentration [ng/µl] | µl available |
|---|---|---|
| DelH F2 | 30 | 25 µl |
| pSB6A1-AraC-lacZ | -11 | 25 µl |
Amplification of DelH F1a
PCR Conditions F1a.W10.A
| Reagent | DelH F1a |
|---|---|
| Template | 0.5 µl of DelH F1a digested Fragment 1:5 (= 8 ng) |
| Primer fw 10 µM | 2.5 µl DelH_f1_PacI_fw |
| Primer rev 10 µM | 2.5 µl DelH_EcoRI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19.5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
Gel does not show any bands.
- => We conclude, that the fragments we expected to be F1a cannot be amplified by PCR. Thus, they were not amplified in the PCR used as template.
PCR Conditions F1a.W10.B
| Reagent | DelH F1a |
|---|---|
| Template | 1 µl ‘‘D. acidovorans’’ glycerol stock |
| Primer fw 10 µM | 2.5 µl DelH_f1_short2_fw |
| Primer rev 10 µM | 2.5 µl DelH_EcoRI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:00 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
See Confirmation of DNA Concentration
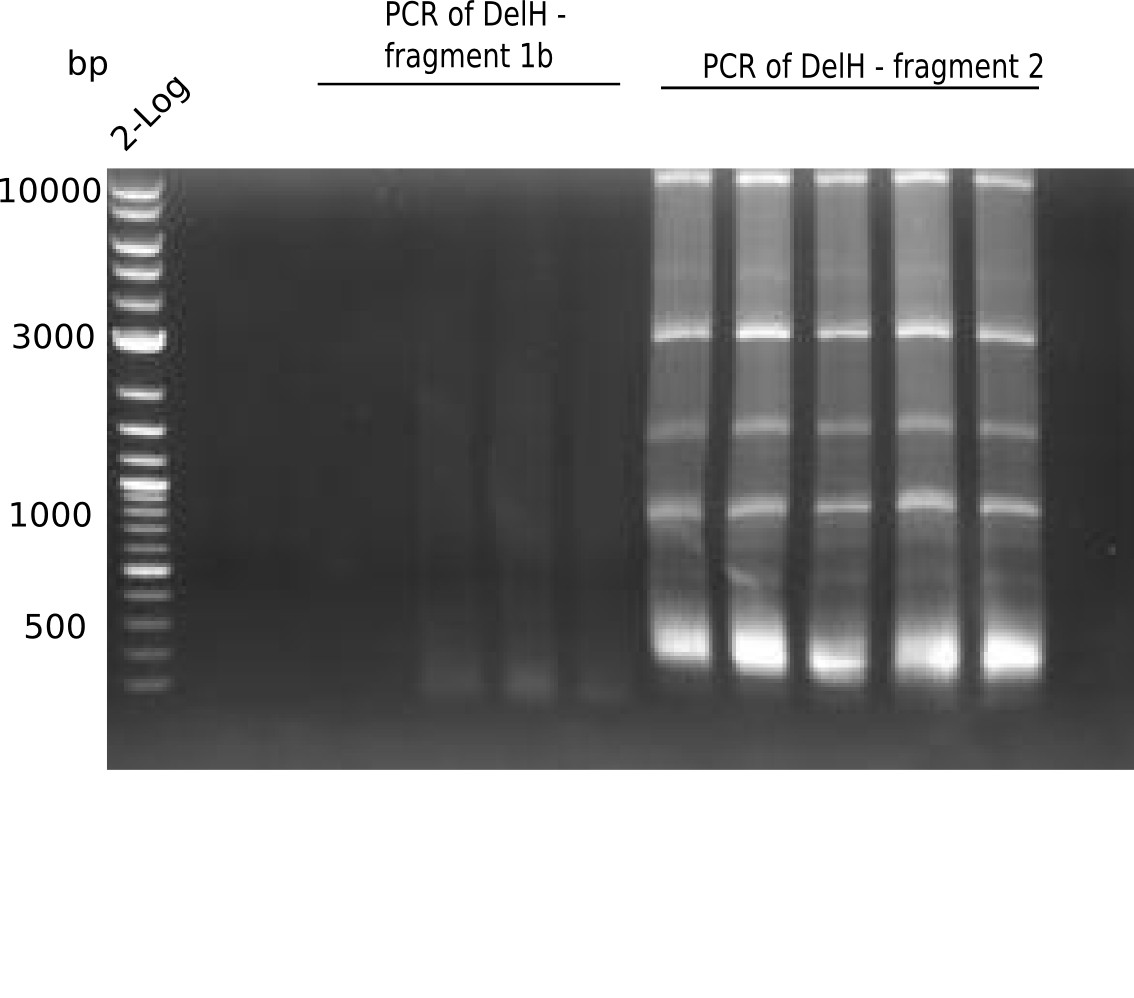
Amplification of DelH F1b
PCR Conditions F1b.W10.C
| Reagent | DelH F1b |
|---|---|
| Template | 1 µl DelH F1b digested Fragment |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
There is no band visible.
- => We conclude, that the fragments we expected to be F1b cannot be amplified by PCR. Thus, they were not amplified in the PCR used as template.
Generation of DelH Plasmid
Confirmation of DNA Concentration
| Fragment | Concentration (by Nanodrop) | µl on gel | µl loading dye | Digested and purified (V + A) | Date (of PCR or Purification) | Check on gel |
|---|---|---|---|---|---|---|
| F1a (PCR) | - | 1 | 9* 1x | - | 03-07-2013 (PCR) | + |
| F1a | 40 | 2 | 9* 1x | V + A | 19-06-2013 | - |
| F1b | 19 | 3 | 1* 6x | V + A | 19-06-2013 | - |
| F1b | 3.5 | 10 | 2* 6x | V + A | 02-07-2013 (V+A) | - |
| F2 | 0.5 | 10 | 2* 6x | V + A | - | |
| F2 | 30.5 | 2 | 10* 1x | V + A (Nucleotide Removal) | 03-07-2013 | + |
| F2 | (1:5 dilution of) 36.9 | 2 | 10* 1x | - | 02-07-2013 | + (BackUP) |
| pSB6A1-AraC-lacZ | 2.2 | 10 | 2* 6x | V + A | 02-07-2013 | + |
| pSB6A1-AraC-lacZ | (1:5 dilution of) 38.5 | 2 | 10* 1x | - | 02-07-2013 | + (BackUP) |
| pSB6A1-AraC-lacZ | -11 | 5 | 1* 6x | V + A (Nucleotide Removal) | 03-07-2013 | + |
DelH F1a (40 ng/µl) was the fragment we used for the first ligation. It is not visible, thus, there was not enough little DNA. Since the screening primer binds to this part of DelH, we could not identify any positive clone.
- => The negative samples (with too little DNA concentration) are discarded.
Amplification of DelH F1a
PCR Conditions F1a.W10.C
| Reagent | DelH F1a |
|---|---|
| Template | 1 µl DelH F1a Fragment 03-07-2013 |
| Primer fw 10 µM | 2.5 µl DelH_Pac_fw |
| Primer rev 10 µM | 2.5 µl DelH_EcoRI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles-PCR | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 66 | 5 | |
| 72 | 120 | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Fragment was gel extracted.
Amplification of DelH F1b
PCR Conditions F1b.W10.D
| Reagent | DelH F1b | DelH F1b |
|---|---|---|
| Template | 1 µl D. acidovorans | 1 µl DelH F1b Fragment (19 ng/µl) |
| Primer fw 10 µM | 2.5 µl DelH_EcoRI_fw | 2.5 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 2.5 µl DelH_f1_SalI_rev | 2.5 µl DelH_f1_SalI_rev |
| Phusion Ready Mix | 25 µl | 25 µl |
| ddH2O | 19 µl | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
No bands visible.
- => Fragment F1b could not be amplified from neither genomic DNA nor PCR fragment. Amplification will be repeated using DMSO in the mix.
PCR Conditions F1b.W10.E
| Reagent | DelH F1b | DelH F1b | DelH F1b | DelH F1b |
|---|---|---|---|---|
| Template | 1 µl D. acidovorans glycerol stock | 1 µl Fragment (19 ng/µl) | 1 µl D. acidovorans glycerol stock | 1 µl PCR Fragment (19 ng/µl) |
| Primer fw 10 µM | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw | 1 µl DelH_EcoRI_fw |
| Primer rev 10 µM | 1 µl DelH_f1_SalI_rev | 1 µl DelH_f1_SalI_rev | 1 µl DelH_f1_SalI_rev | 1 µl DelH_f1_SalI_rev |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl | 6 µl | 6 µl |
| DMSO | - | - | 1 µl | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 2:15 min | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 2:15 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
- Using hot start 98°C
Result
Expected band: 5 Kb
Only PCR of fragment 19 ng/µl + DMSO was successful. Maybe there is another pale band at the second PCR of DelH F1b.
- => Repeat both PCRs.
Result
PCR using conditions F1b.W10.E was repeated.
Expected band: 5 Kb,Entire PCR mix was loaded.
PCR did not result in any fragments. Interestingly, both older PRCs resulted in a fragment at ~5 Kb and other lines, but the fragment from the genomic DNA is slightly larger.
- => Both fragments were cut and gel isolated, check correct length of F1b!
- => Perform PCR from the PCR fragment obtained from the genomic DNA.
Amplification of DelH F2
PCR Conditions F2.W10.C
- 2x 50 µl
| Reagent | DelH F2 |
|---|---|
| Template | 1 µl Fragment "BackUP" |
| Primer fw 10 µM | 2.5 µl DelH_f2_SalI_fw |
| Primer rev 10 µM | 2.5 µl DelH_f2_KpnI_rev |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 8 Kb
Fragment 2 was successfully amplified.
- => It was cut and gel purified.
Preparation of new Template DNA
- D. acidovorans glycerol stock was streaked onto LB plates and incubated ON at 37°C.
- D. acidovorans did not grow, since LB media is not appropriate for them. Prepare new ACM media and plates and streak D. acidovorans.
Amplification of Backbone
PCR Conditions BB.W10.D
- 2x 50 µl
| Reagent | Backbone |
|---|---|
| Template | 1 µl Fragment "BackUP" |
| Primer fw 10 µM | 2.5 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 2.5 µl AraCbbPacI_rev2 |
| Phusion Flash Ready Mix | 25 µl |
| ddH2O | 19 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 7.3 Kb
No band visible.
- => Repeat using DMSO.
PCR Conditions BB.W10.E
| Reagent | Backbone | Backbone | Backbone |
|---|---|---|---|
| Template | 1 µl of BackUP | 1 µl V+A 2,2 ng/µl template | 1 µl V+A 2,2 ng/µl template |
| Primer fw 10 µM | 1 µl AraCbb_KpnI_fw | 1 µl AraCbb_KpnI_fw | 1 µl AraCbb_KpnI_fw |
| Primer rev 10 µM | 1 µl AraCbbPacI_rev2 | 1 µl AraCbbPacI_rev2 | 1 µl AraCbbPacI_rev2 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl | 6 µl |
| DMSO | - | - | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 2:30 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 2:30 min | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
- Using hot start (in new cycler)
Result
Expected band: 7.3 Kb
Gel does not show the expected fragment.
- => Next PCR for the Backbone should be done with longer elongation time and variations: think about using the a new colony of plate.
PCR Conditions BB.W10.F
| Reagent | Backbone | Backbone | Backbone | Backbone |
|---|---|---|---|---|
| Template | 1 µl Fragment 24 ng/µl | 1 µl of Fragment pSB6A1 gel-purified 3-6 89 ng/µl | 1 µl of digested Fragment 3-7 -11 ng/µl | 1 µl of purified Fragment Isoprop 2.2 ng/µl |
| Primer fw 10 µM | 1 µl DelH_f2_SalI_fw | 1 µl AraCbb_KpnI_fw | 1 µl DelH_f2_SalI_fw | 1 µl AraCbb_KpnI_fw 1 |
| Primer rev 10 µM | 1 µl DelH_f2_KpnI_rev | 1 µl AraCbbPacI_rev2 | 1 µl DelH_f2_KpnI_rev | 1 µl AraCbbPacI_rev2 |
| Phusion Flash Ready Mix | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 7 µl | 7 µl | 7 µl | 7 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 66 decrease by 0.5 | 5 | |
| 72 | 180 | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 180 | |
| 1 | 72 | 7 min |
| 1 | 4 | inf |
Result
Expected band: 7.3 Kb,Entire PCR mix loaded (pSB6A1-AraC-lacZ)
Only PCRs from Fragment 24 ng/µl and Fragment 3-6 produced a nice fragment.
- => Bands were cut and gel isolated.
- => PCR amplification from restriction digested fragment did not work! This is unexpected, because actually, it should. Maybe exclude the previous restriction digested fragments "V+A" from ligation.
Preparation of new Template DNA
- psB6A1-LacZ-AraC: 2 ml LB Amp were inocculated from the plate and incubated ON at 28°C (all other shakers occupied).
- Mini prep was performed.
- 1 µl mini prep DNA was electroporated into E.coli DH10ß according to Electroporation of E. coli DH10β
 "
"