Team:Heidelberg/Templates/DelH week19
From 2013.igem.org
Contents |
02-09 - 08-09-13
Characterization of DelH Plasmid pHM04 30-08 Clone 12
Maxiprep
- 1 ml of an ON culture DH10ß-DelH were inocculated in 200 ml LB Amp and cultured ON at 37°C.
- Maxi prep was performed by upscaling of mini precipitation protocol.
Result
The maxi prep yielded 6x 180 µl.
| Maxi prep | Concentration [µg/µl] |
|---|---|
| 1 | 5.40 |
| 2 | 5.37 |
| 3 | 5.42 |
| 4 | 5.40 |
| 5 | 5.38 |
| 6 | 5.42 |
- Sequencing showed a mutation at the beginning of DelH
- Next steps
- => Sequencing the other 3 positive colonies => hopefully one of them has no mutation at the beginning of DelH
- => Sequence D. acidovorans, maybe the organism SPH1 was not perfectly sequenced => amplify with FS35 & DN07
Characterization of DelH Plasmid pHM04 30-08 Clones 4, 6, 15
Sequencing
- Miniprep performed
- Send in for sequencing => no reliable result
- Maxiprep performed => Clone 4: c= 5.51 µg/µl, Clone 6: c= 3.71µg/µl, Clone 15: c= 4.65 µg/µl
- Send in for sequencing with DN07 again
Result
| Sample | Alignment File |
|---|---|
| Midi-col 4 | File:Heidelberg Sequencing Result pHM04 DN07 colony 04.clustal.txt |
| Midi-col 6 | sequencing insufficient |
| Maxi-col 15 | sequencing insufficient |
Electroporation
Because the sequenced DelH colony 12 has a mutation at the beginning of the coding DNA, we are sequencing the other colonies and parallel we transformed with the rest of the same Gibson Mix (15 µl) used for the electroporation before (colonies 4, 6, 12, 15). In advance, we purified it via isoprop purification protocol.
| Sample | Isoprop purified Gibson Mix | Colonies grown? |
|---|---|---|
| A | 10 µl | yes |
| B | 20 µl | yes (a lot more than on sample A) |
Colony-PCR conditions CP.W19.A
There were 30 colonies screened. 3 per tube and in total 10 colonies of sample A & 20 colonies of sample B
| Template | 3 PC of S-A | 3 PC of S-A | 4 PC of S-A | 3 PC of S-B | 3 PC of S-B | 4 PC of S-B | 3 PC of S-B | 3 PC of S-B | 4 PC of S-B |
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 1-3 | 4-6 | 7-10 | 11-13 | 14-16 | 17-20 | 21-23 | 24-26 | 27-30 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 |
| iTaq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 60 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 60 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
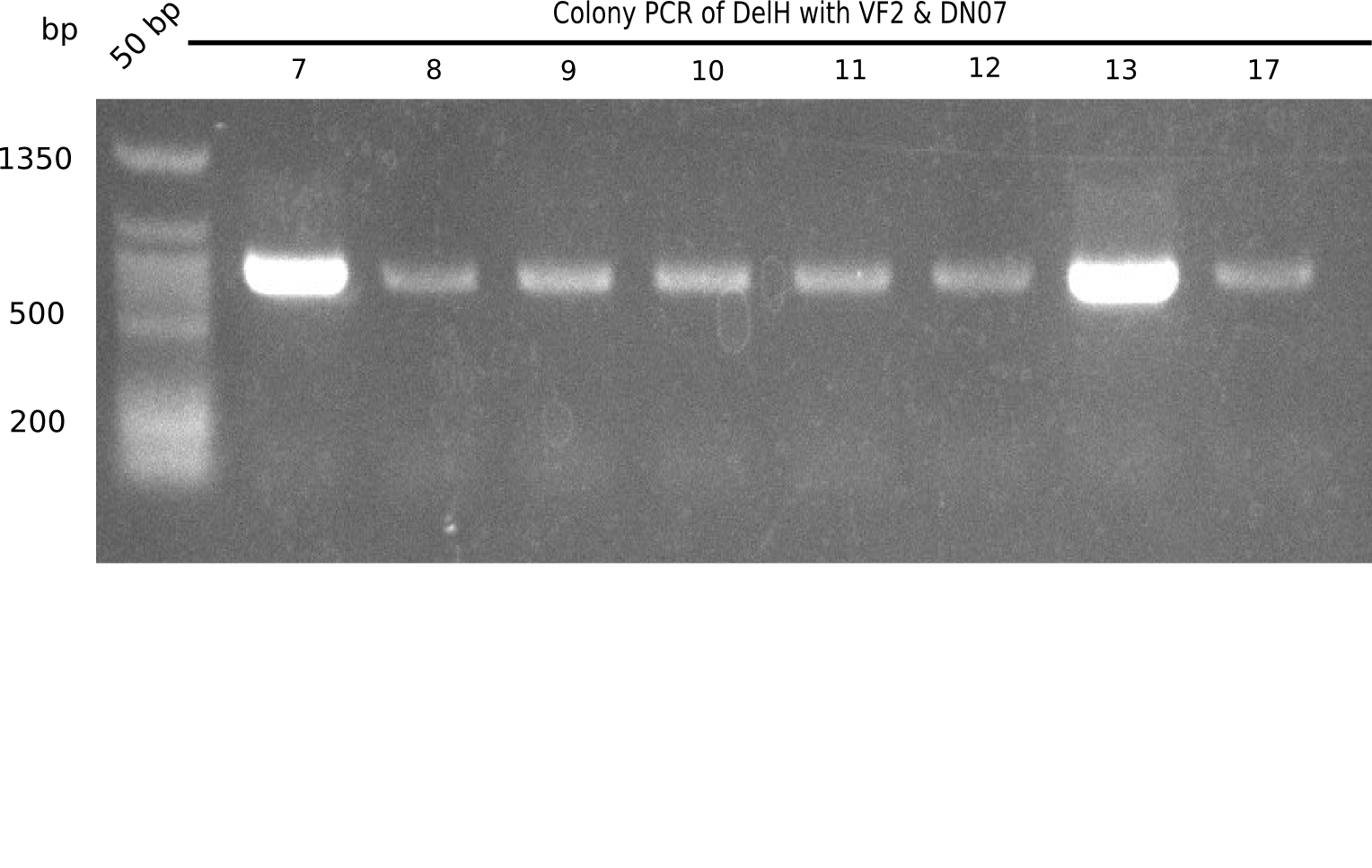
Result
Expected band: 663 bp
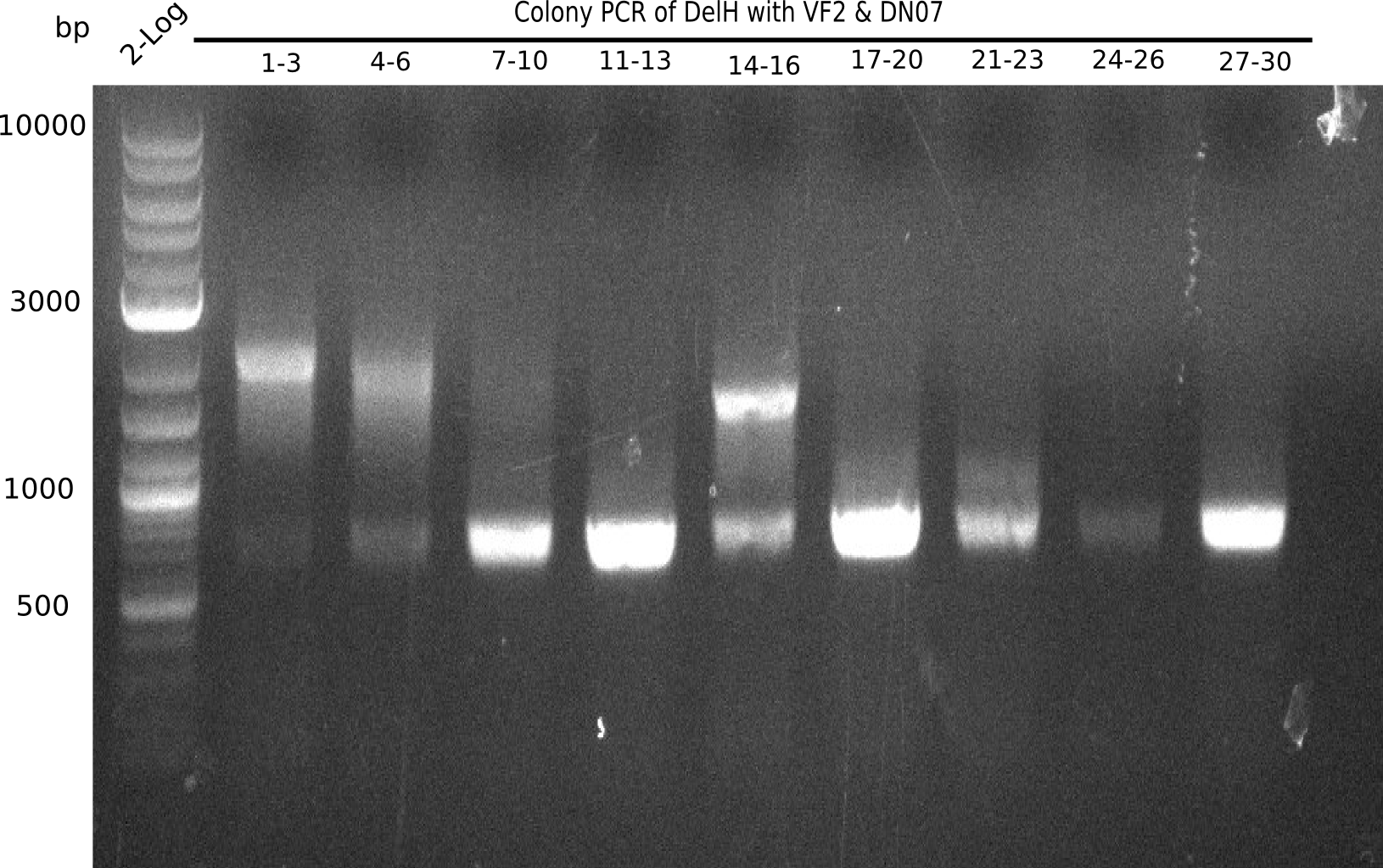
l1:2log, l2:colonies 1-3, l3:colonies 4-6, l4:colonies 7-10, l5:colonies 11-13, l6:colonies 14-16, l7:colonies 17-20, l8:colonies 21-23, l9:colonies 24-26, l10:colonies 27-30
l4, l5,l8,l10:show the specific band at 663 bp
Some of the colony mixes show positive band.
- => Screen again colonies 7-10, 11-13, 17-20, 27-30 in a single PCR tubes.
Colony-PCR conditions CP.W19.B
| Template | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 17 | 18 | 19 | 20 | 27 | 28 | 29 | 30 |
| Expected length [bp] | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 | 663 |
| Named | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 17 | 18 | 19 | 20 | 27 | 28 | 29 | 30 |
| Primer fw 10 µM | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 | 2 µl VF2 |
| Primer rev 10 µM | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 | 2 µl DN07 |
| iTaq Polymerase (2x) | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl | 10 µl |
| ddH2O | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl | 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
Colonies 7,13, 19 & 29 show a definit result
- => Add medium and perform a mediprep the next day for sending in for sequencing.
Midiprep
Performed with 300 µl P1, 600 µl P2 and after incubating for 180 450 µl S3. Protocol was performed with isoprop/ethanol purification.
| Sample | Concentration [ng/µl] |
|---|---|
| Midi-col 7 | 2628.5 |
| Midi-col 13 | 2379.3 |
| Midi-col 19 | 2846.6 |
| Midi-col 29 | 2846.6 |
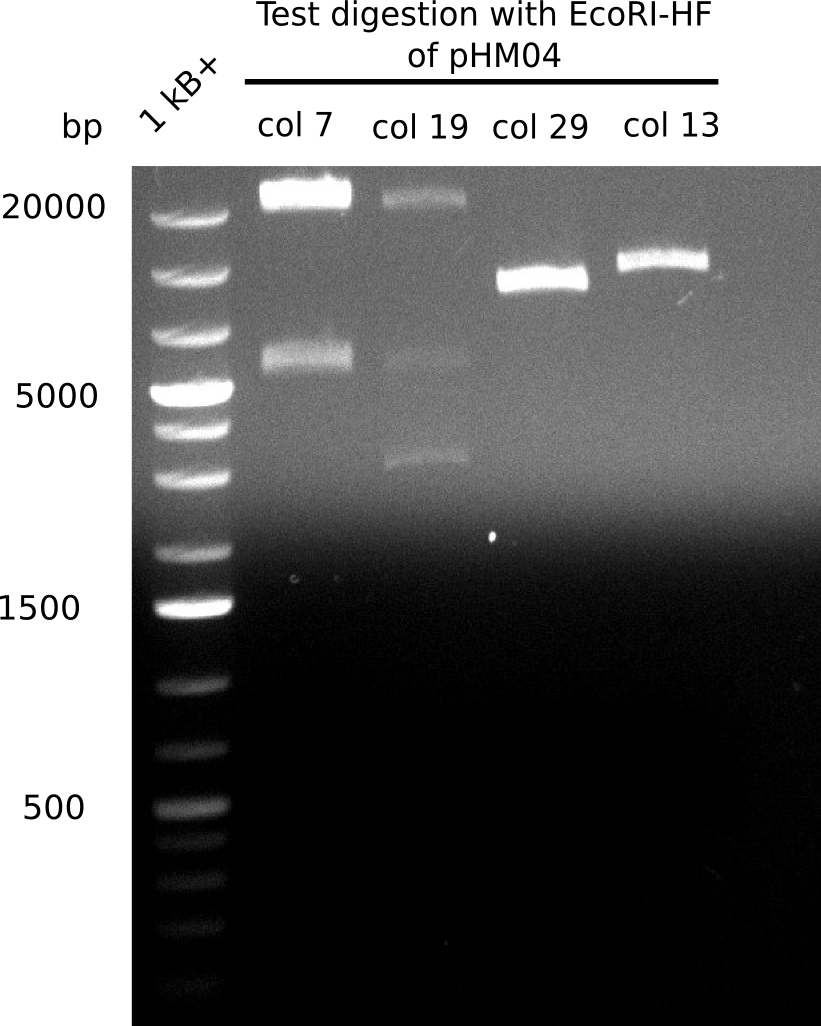
Test Restriction Digest
We performed a test digest with EcoRI-HF of the screened samples col 7, col 13, col 19, col 29. We expect a band of 17.9 Kb and 4.9 Kb.
| Sample | Concentration [ng/µl] | Amount in restriction digest | EcoRI-HF [µl] | CutSmartBuffer [µl] | ddH2O [µl] | Total amount [µl] |
|---|---|---|---|---|---|---|
| Midi-col 7 | 2628.5 | 0.5 µl = 1314.25 ng | 1 | 2 | 16.5 | 20 |
| Midi-col 13 | 2379.3 | 0.5 µl = 1189.65 ng | 1 | 2 | 16.5 | 20 |
| Midi-col 19 | 2846.6 | 0.5 µl = 1423.3 ng | 1 | 2 | 16.5 | 20 |
| Midi-col 29 | 2846.6 | 0.5 µl = 1423.3 ng | 1 | 2 | 16.5 | 20 |
- Incubation time: 1 h at 37 °C
Result
Expected bands: 17.9 Kb and 4.9 Kb
Colonies 7 & 13 show a definit result
- => Add medium and perform a mediprep the next day for send in for sequencing.
Sequencing
The colonies 7 & 19 were sent in MWG for sequencing. There for we prepared 15 µl of the plasmid (midiprep) with a concentration of 50-100 ng/µl and added 2 µl DN07 primer (10µM)
| Sample | Concentration [ng/µl] | Amount for sequencing | ddH2O added up to 15 µl | Final concentration [ng/µl] |
|---|---|---|---|---|
| Midi-col 7 | 2628.5 | 0.5 µl = 1314.25 ng | 14.5 | 87.62 |
| Midi-col 19 | 2846.6 | 0.5 µl = 1423.3 ng | 14.5 | 94.89 |
| Maxi-col 4 | 5500 | 2 µl of 1:10 dilution = 1100 ng | 13 | 73.3 |
Result
| Sample | Alignment File |
|---|---|
| Midi-col 7 | File:Heidelberg Sequencing Result pHM04 DN07 colony 7.clustal.txt |
| Midi-col 19 | File:Heidelberg Sequencing Result pHM04 DN07 colony 19-2.clustal.txt |
| Maxi-col 4 | File:Heidelberg Sequencing Result pHM04 DN07 colony 04.clustal.txt |
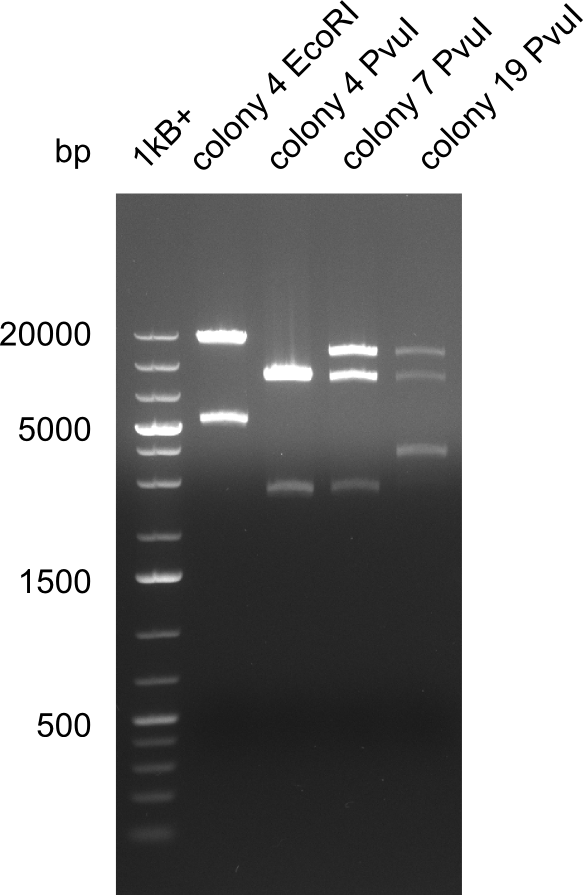
Test Restriction Digest
We performed a test digest with EcoRI-HF and PvuI-HF of the screened samples col 4, col 7, col 19. We expect a band of 17.9 Kb and 4.9 Kb for the digest with EcoRI-HF and bands of 11.5 Kb, 8.5 Kb and 2.6 Kb.
| Sample | Concentration [ng/µl] | Amount in restriction digest | Enzyme | CutSmartBuffer [µl] | ddH2O [µl] | Total amount [µl] |
|---|---|---|---|---|---|---|
| Midi-col 4 | 5500 | 0.25 µl = 1375 ng | EcoRI-HF 1 µl | 2 | 16.75 | 20 |
| Midi-col 4 | 5500 | 0.25 µl = 1375 ng | PvuI-HF 1 µl | 2 | 16.75 | 20 |
| Midi-col 7 | 2628.5 | 0.5 µl = 1314.25 ng | PvuI-HF 1 µl | 2 | 16.5 | 20 |
| Midi-col 19 | 2846.6 | 0.5 µl = 1423.3 ng | PvuI-HF 1 µl | 2 | 16.5 | 20 |
- Incubation time: 1:20 h at 37 °C
Result
Expected band: 17.9 Kb and 4.9 Kb (EcoRI-HF) and 11.5 Kb, 8.5 Kb and 2.6 Kb (PvuI)
Colony 7 is the only colony which is positiv for all restriction digests.
- => Electroporate plasmid together with DelRest plasmid, as well as the Methylmanolyl-CoA plasmid.
Sequencing of D. acidovorans
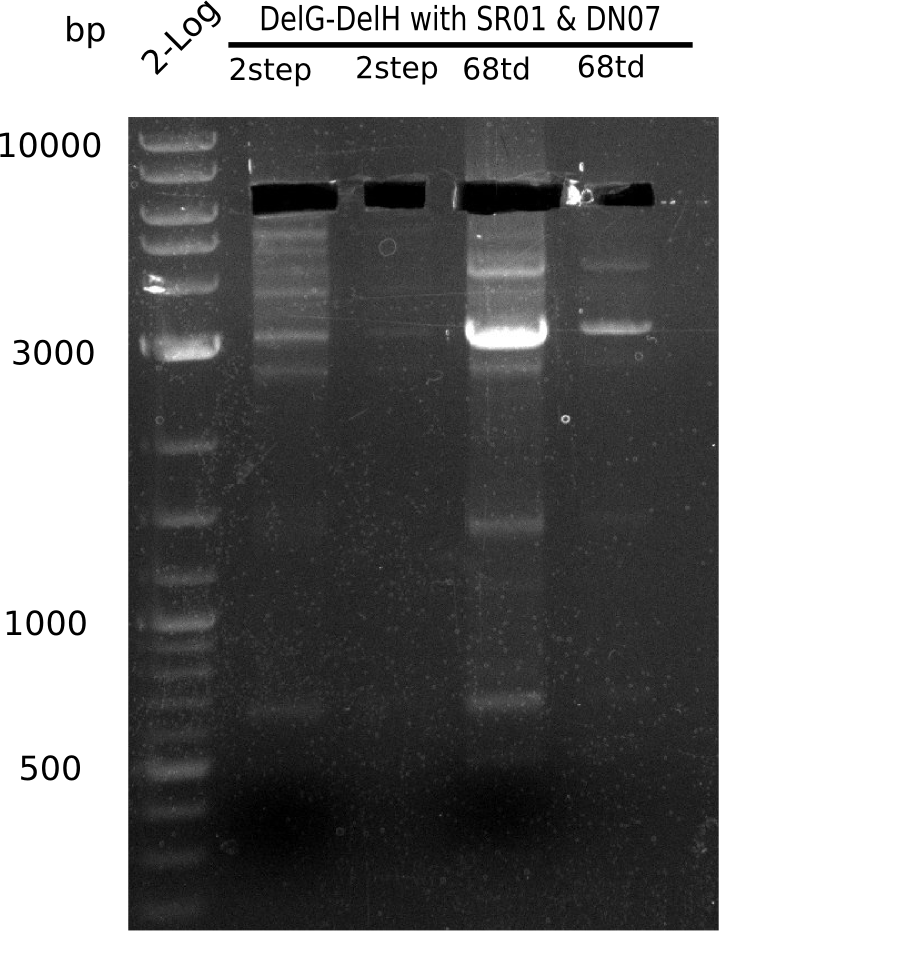
PCR Conditions DA.W19.A
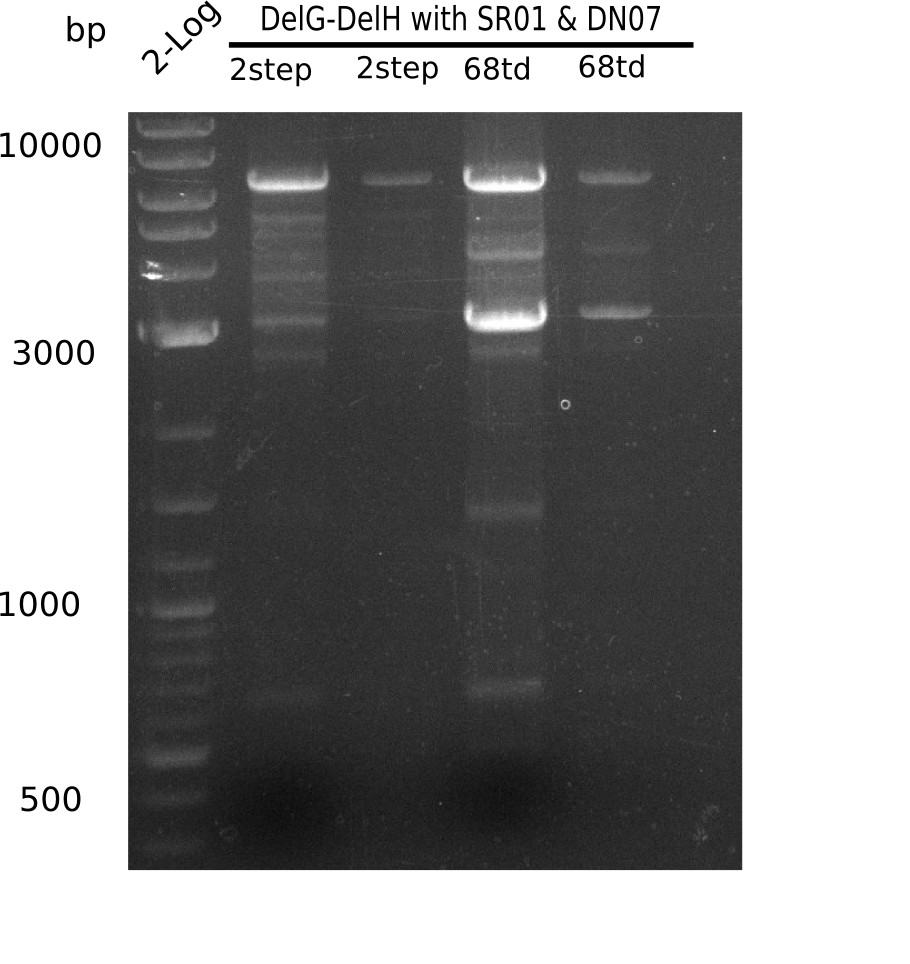
For being sure that the mutation at the beginning of DelH is not some mistake in the sequenced genome we amplify this part with one primer binding the delG (FS35) and the screening primer binding at the beginning of DelH (DN07) and let it sequence with GATC.
| Template | 1 µl glycerol stock of D. acidovorans | 1 µl glycerol stock of D. acidovorans |
|---|---|---|
| Expected length [Kb] | 6.5 | 6.5 |
| Named | TD | 2step |
| Primer fw 10 µM | 2 µl FS35 | 2 µl FS35 |
| Primer rev 10 µM | 2 µl DN07 | 2 µl DN07 |
| Phusion Flash (2x) | 10 µl | 10 µl |
| ddH2O | 4 µl | 4 µl |
| DMSO | 1 µl | 1 µl |
| Cycles | Temperature A [°C] | Time [s] | Temperature B[°C] | Time [s] |
|---|---|---|---|---|
| 1 | 98 | 10 | 98 | 10 |
| 12 | 98 | 1 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | - | - | |
| 72 | 130 | 72 | 120 | |
| 18 | 98 | 1 | 98 | 1 |
| 66 | 5 | - | - | |
| 72 | 130 | 72 | 120 | |
| 1 | 72 | 10 min | 72 | 10 min |
| 1 | 12 | inf | 12 | inf |
Result
Expected band: 6.5 Kb
Gel shows expected band.
- => Fragments were cut but not extracted, because side bands were cut too.
- => Run again PCR with 72.3 °C annealing temperature.
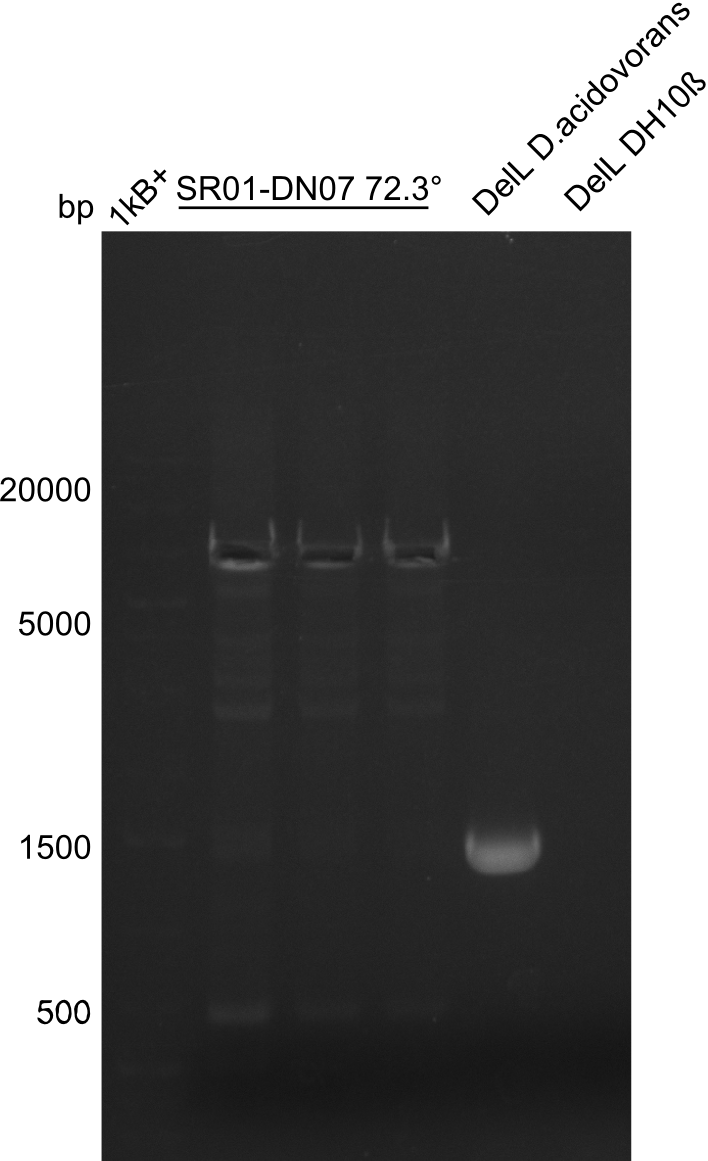
PCR Conditions DA.W19.B
| Template | 1 µl glycerol stock of D. acidovorans |
|---|---|
| Expected length [Kb] | 6.5 |
| Named | FS35-DN07 |
| Primer fw 10 µM | 2 µl FS35 |
| Primer rev 10 µM | 2 µl DN07 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 4 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Result
Expected band: 6.5 Kb
Gel shows expected band at ~7 Kb.
- => The fragment was cut and gel purified. Nevertheless the bands were rather weak, so the PCR was repeated.
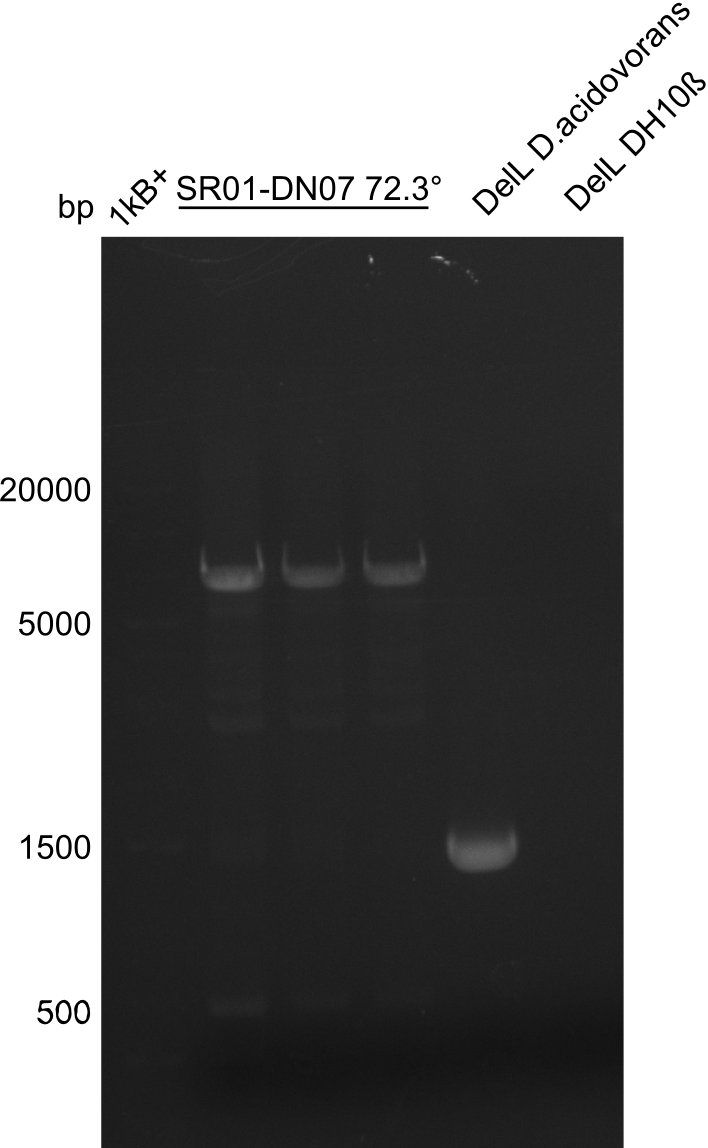
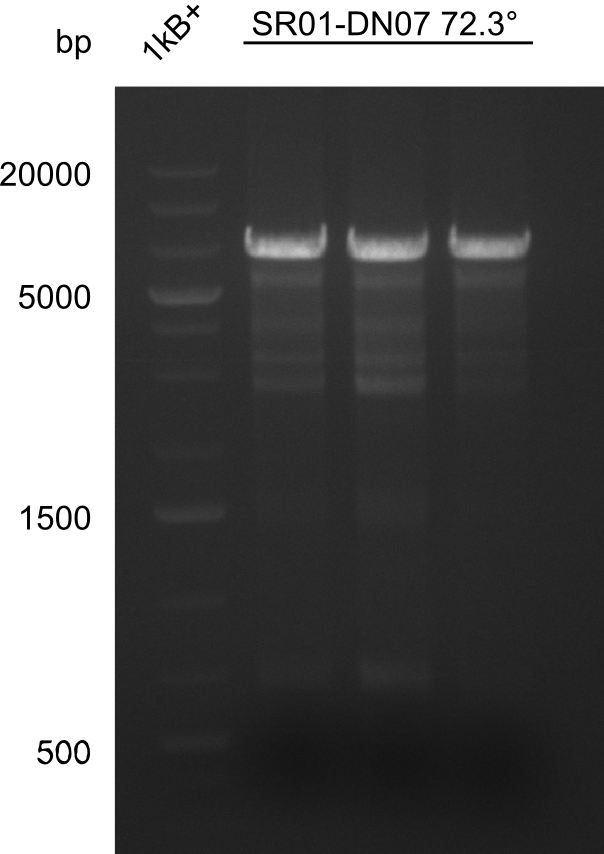
PCR Conditions DA.W19.C
| Template | 1 µl glycerol stock of D. acidovorans |
|---|---|
| Expected length [Kb] | 6.5 |
| Named | FS35-DN07 |
| Primer fw 10 µM | 2 µl FS35 |
| Primer rev 10 µM | 2 µl DN07 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 4 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 72.3 | 5 | |
| 72 | 120 | |
| 1 | 72 | 10 min |
| 1 | 12 | inf |
Result
Expected band: 6.5 Kb
Gel shows expected fragment.
- => The fragment was cut and gel purified.
- => Send in for sequencing with DN07.
 "
"