Team:Heidelberg/Templates/DelH week21
From 2013.igem.org
16-09 - 22-09-13
Generation of DelH Plasmid pHM04 15-09
Colony-PCR Conditions CP.W21.A
| Template | 96x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12H |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| iTaq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature DelH [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
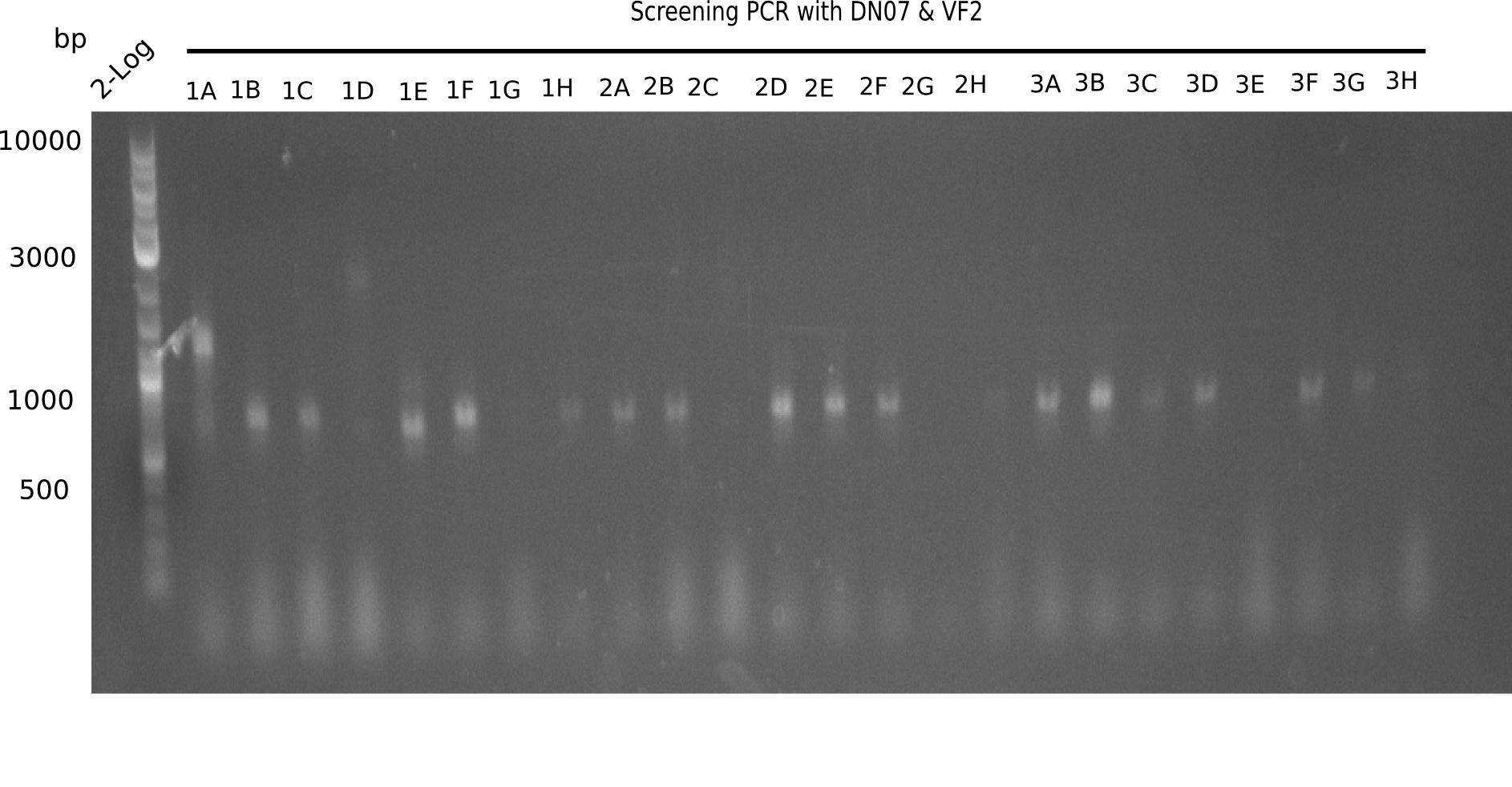
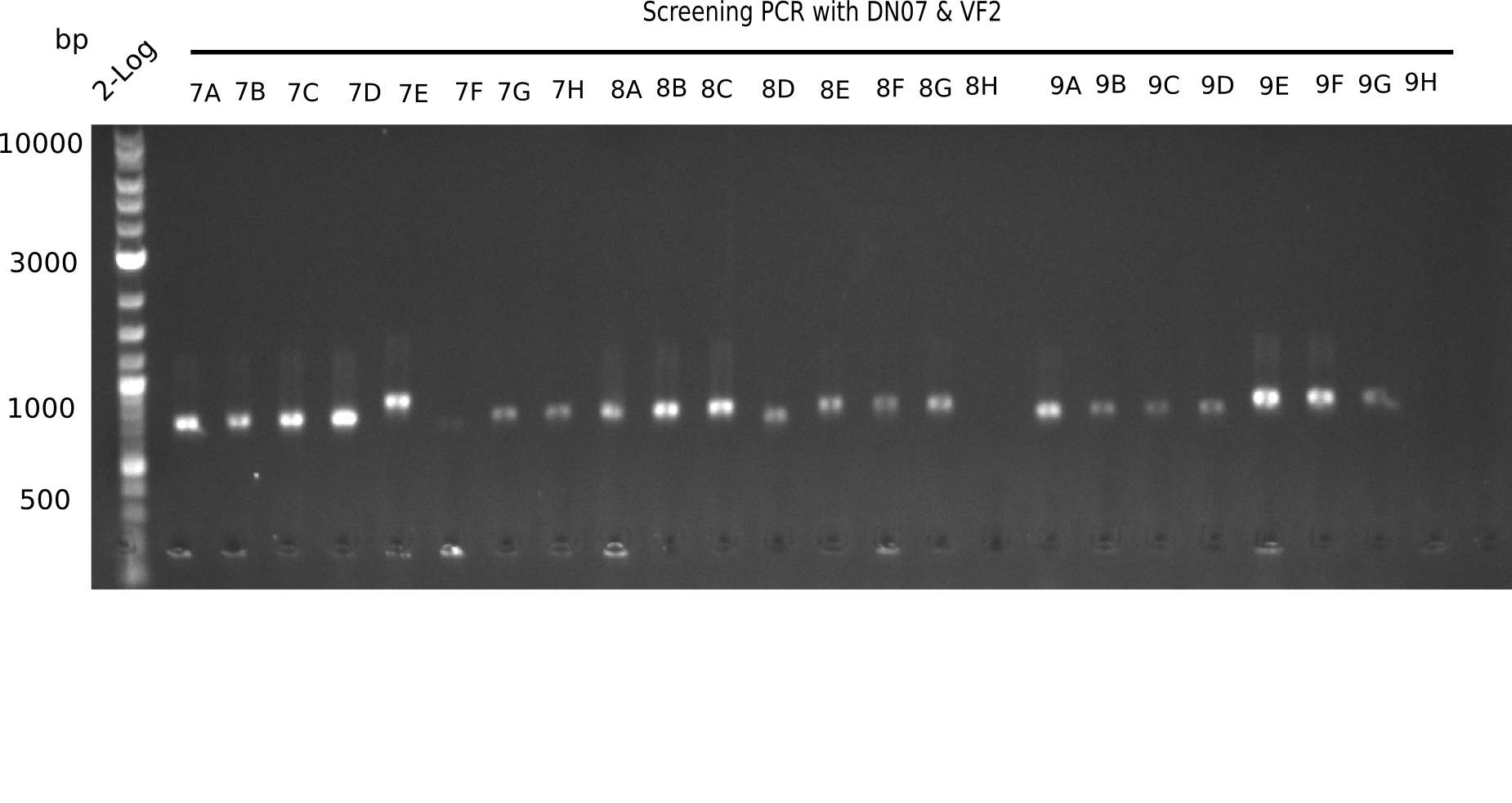
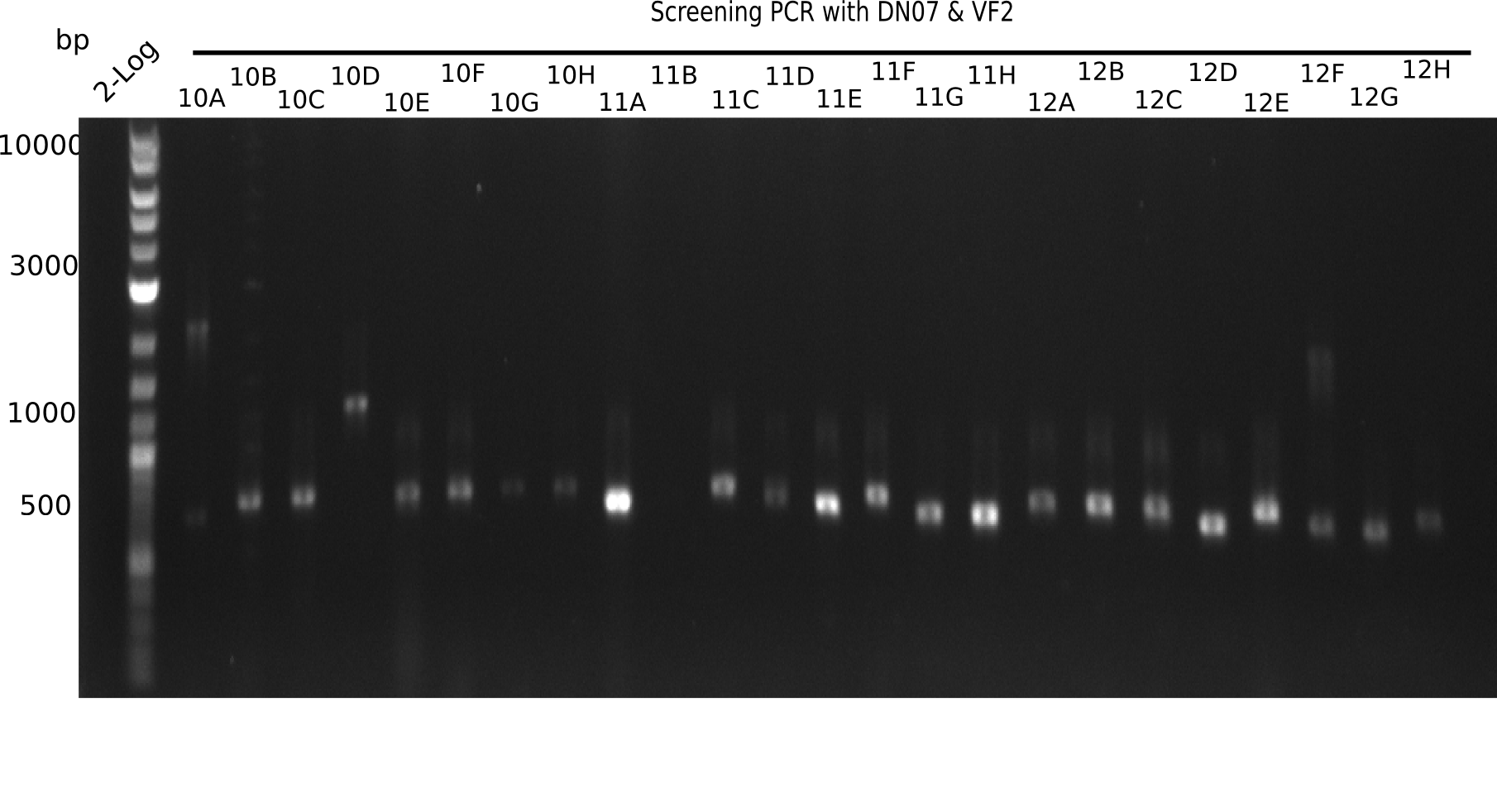
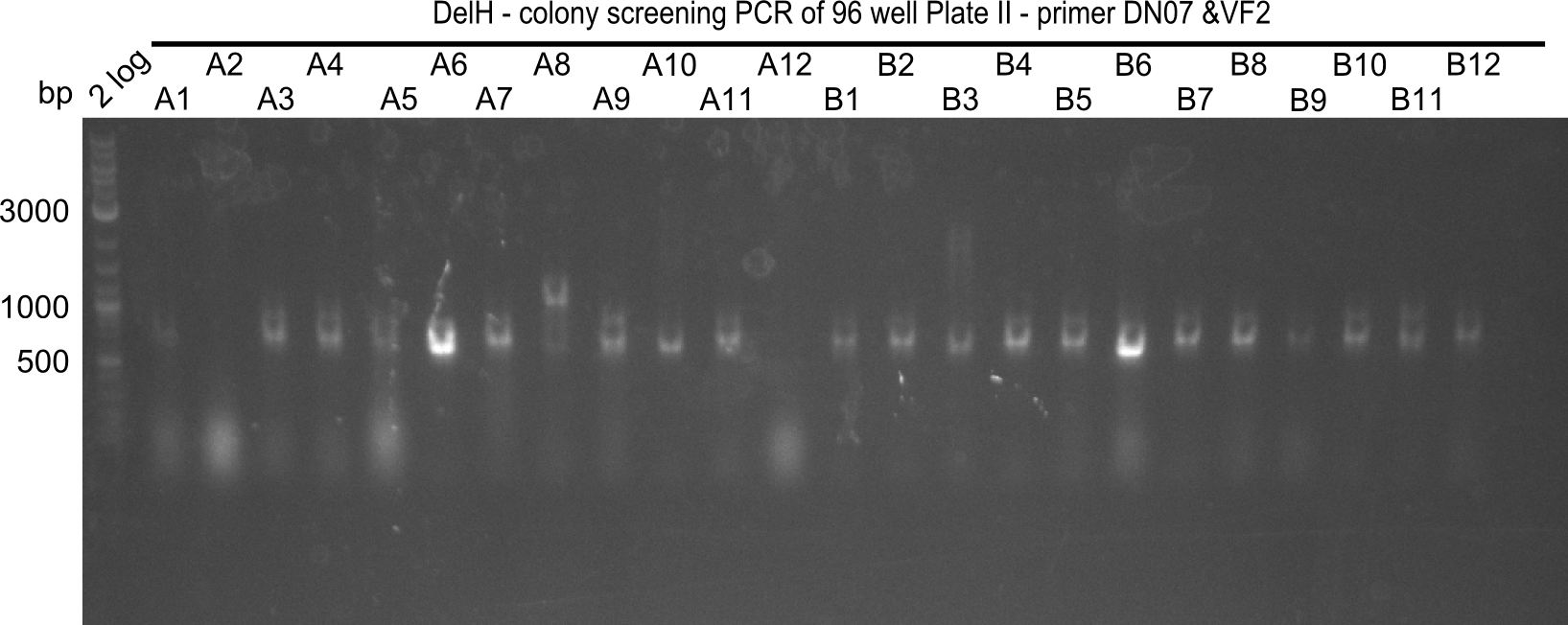
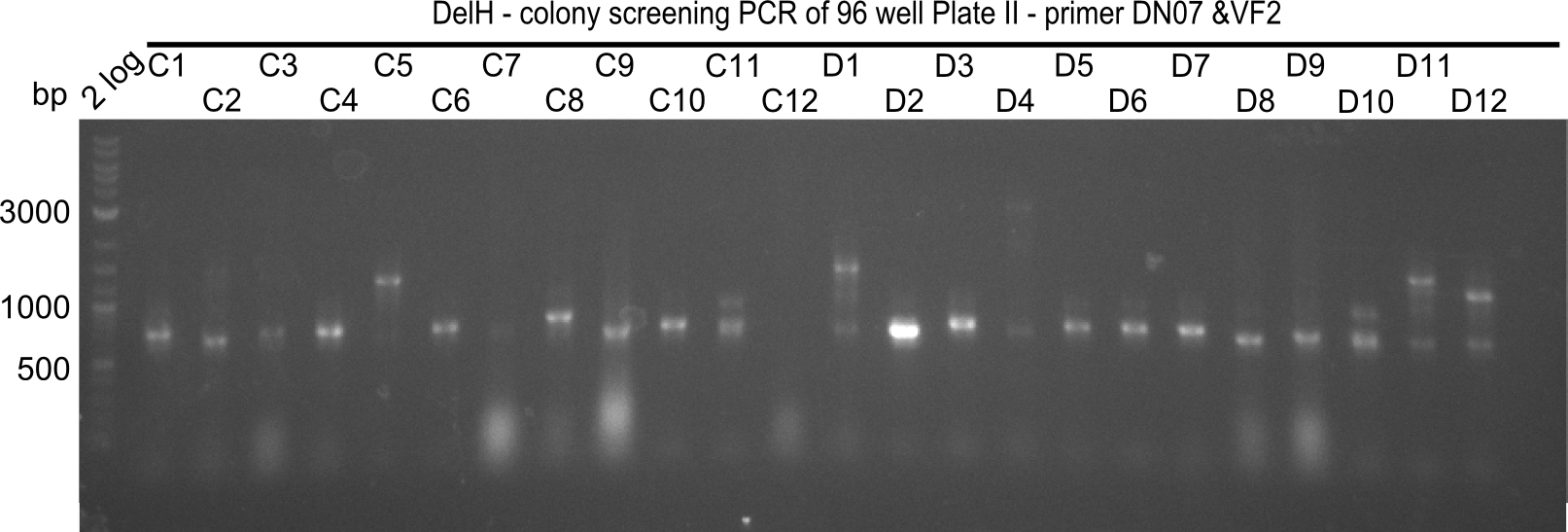
Result
Expected band: 663 bp
The following colonies seem positive, because they show the expected band at 663 bp.
| Picture | Positive colonies |
|---|---|
| 21.1 | 1B, 1C, 1E, 1F, 1H, 2A, 2B, 2D, 2E, 2F, 3A, 3B, 3C, 3D, 3F, 3G |
| 21.2 | 4A, 4C, 4D, 4G, 5A, 5B, 5C, 5D, 5E, 6A, 6B, 6C, 6E, 6F |
| 22.3 | 7A, 7B, 7C, 7D, 8A, 8B, 8C, 9A, 9E, 9F |
| 22.4 | 11 A, 11E, 11G, 11H, 12D, 12E |
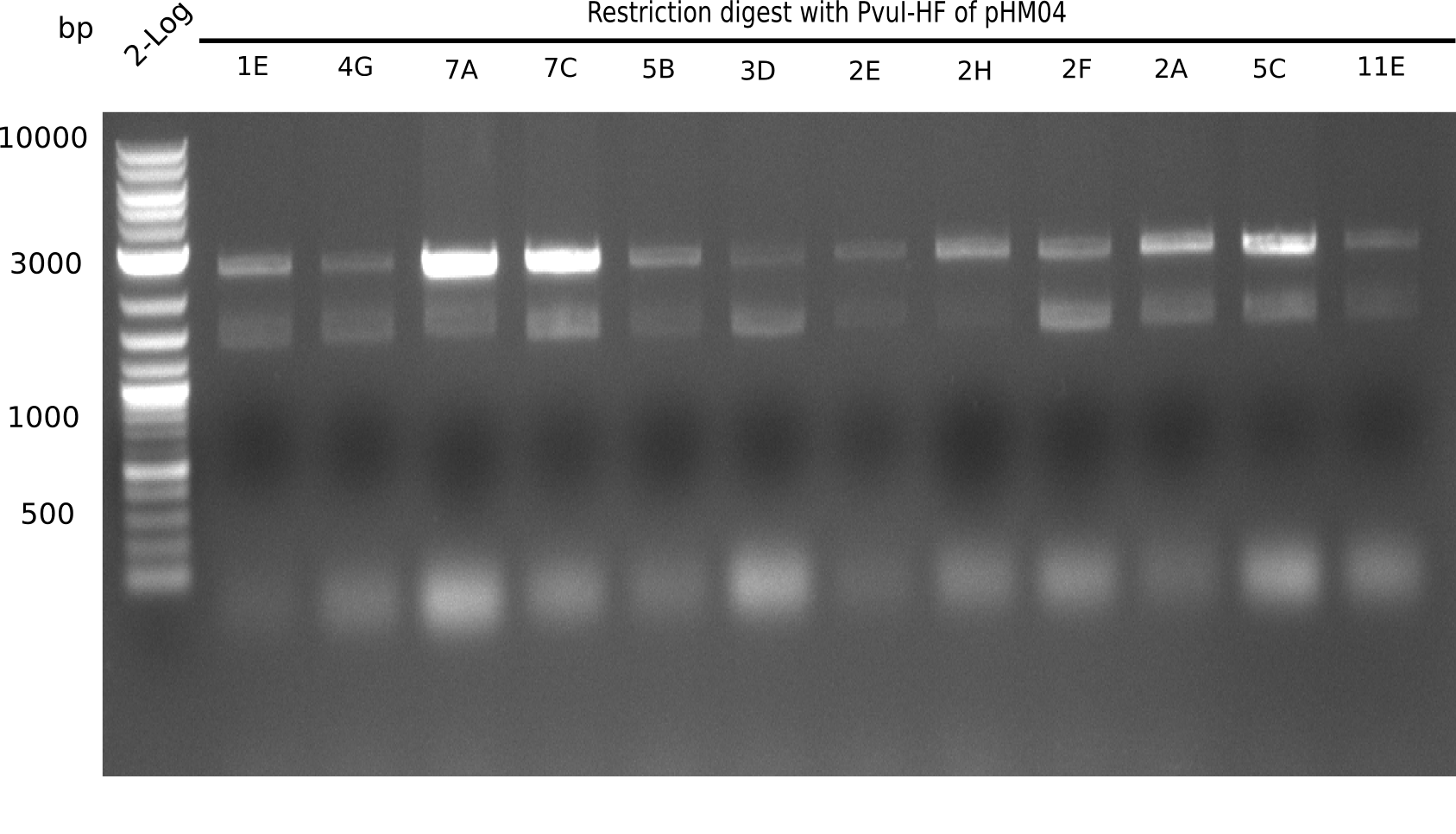
Test Restriction Digest
24 of these positive screened colonies were minipreped and digested with PvuI
| Sample | CutSmartBuffer | PvuI |
|---|---|---|
| 8.5 µl of colonies | 1 µl | 0.5 µl |
Another restriction digest was performed. Colony 7D was digested with EcoRI-HF
| Sample | CutSmartBuffer | EcoRI-HF |
|---|---|---|
| 8.5 µl of colony 7D | 1 µl | 0.5 µl |
Result
Expected bands: 11,621, 8,690 and 2,685 bp (PvuI) and 17,801 and 5,105 bp (EcoRI)
None of the analyzed clones shows correct band pattern.
- => Clones are discarded.
Amplification of Backbone
New Template
Because the backbone amplification yield from pSB6A1 + BBa_J04450 from Spring distribution 2012 (plate 1, well K1) was so low and the screening did not result in positive restriction digests with PvuI or EcoRI, we go a step back and transform E.coli ToP10 with a new pSB6A1 part from the 2013 distribution. Therefore, we transform on the one hand with the part on plate 2 well 2L and on the other hand with the part from the plate 5 well 1K from the Spring 1023 diestricbution.
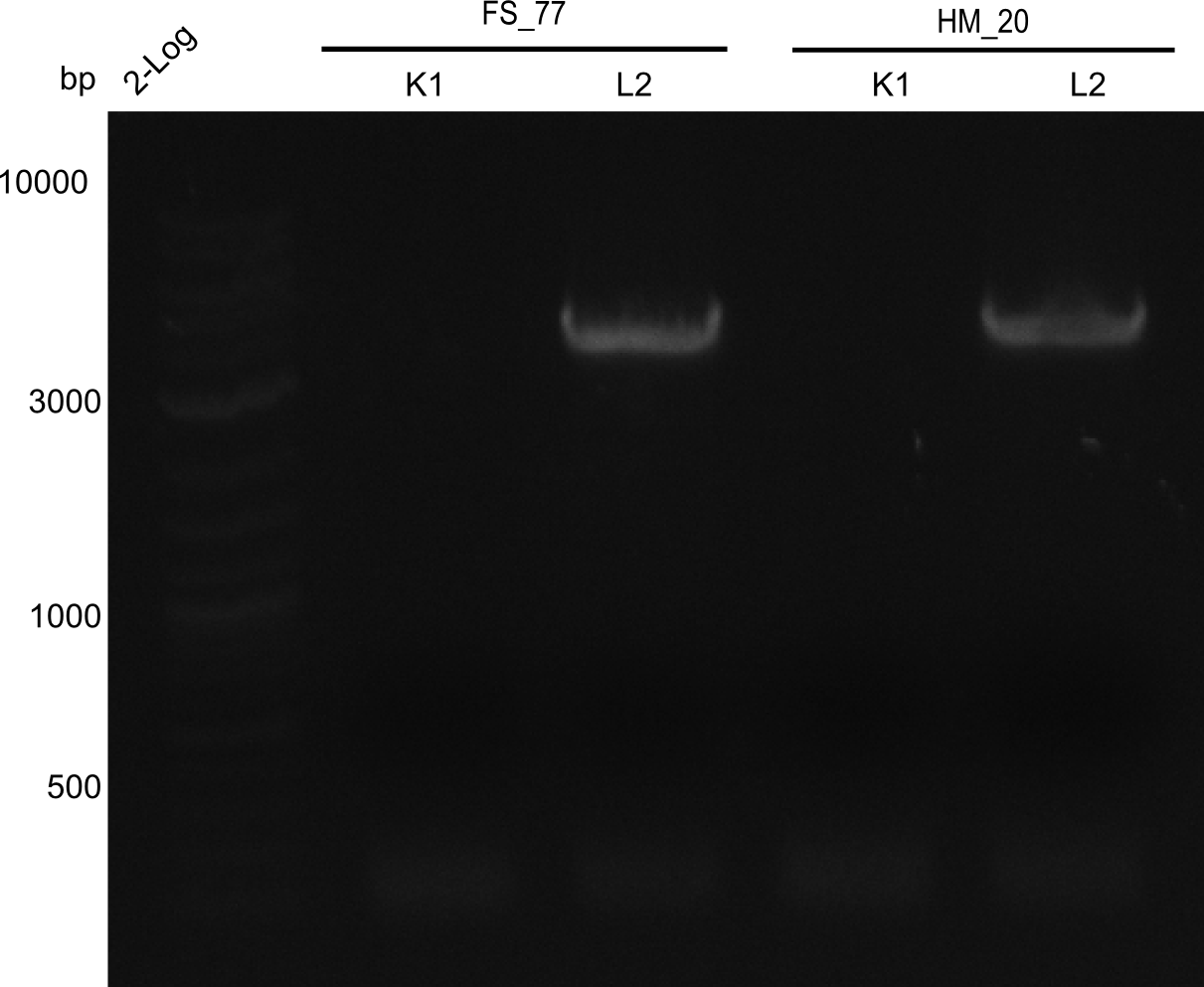
Aditionally we ran a PCR directly from the two wells with the backbone primers (HM20, HM17 & FS77).
PCR Conditions BB.W21.A
Amplification of pSB6A1 directly from the parts registry
1x PCR with HM_20 and 1x PCR with FS_77
| Reagent | BB pSB6A1 | BB pSB6A1 | BB pSB6A1 | BB pSB6A1 |
|---|---|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 | 4.4 | 4.4 |
| Template | 1 µl pSB6A1 plate 5, well K1 | 1 µl pSB6A plate 2, well L2 | 1 µl pSB6A1 plate 5, well K1 | 1 µl pSB6A plate 2, well L2 |
| Primer 10 µM fw | 1 µl HM_17 | 1 µl HM_17 | 1 µl HM_17 | 1 µl HM_17 |
| Primer 10 µM rev | 1 µl FS_77 | 1 µl FS_77 | 1 µl HM_20 | 1 µl HM_20 |
| Phusion Flash Master Mix (2x) | 10 | 10 | 10 | 10 |
| DMSO | 1 | 1 | 1 | 1 |
| ddH2O | 6 | 6 | 6 | 6 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
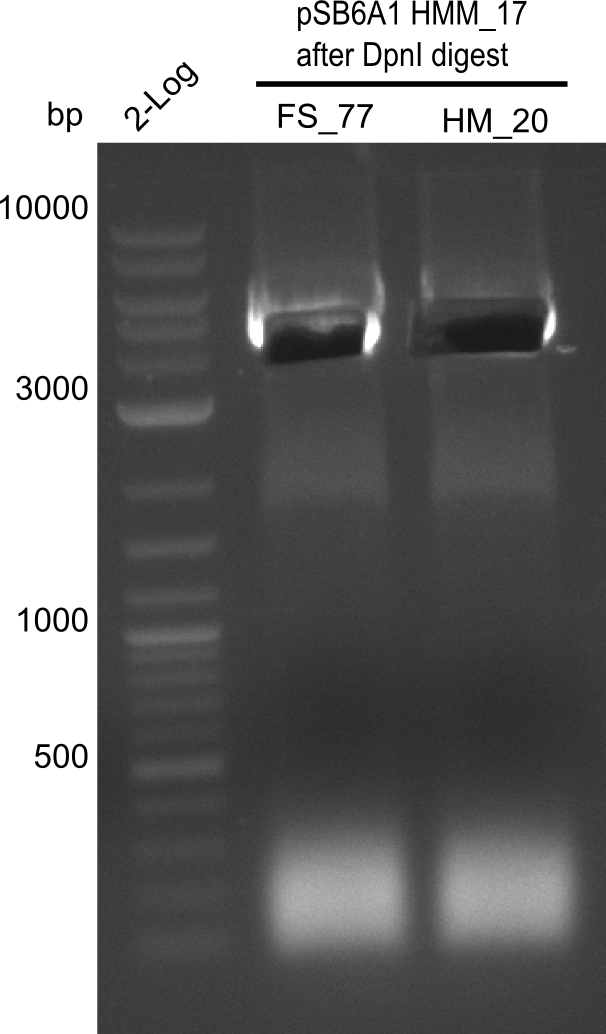
Result
Expected band: 4. Kb
Amplification worked with the template pSB6A1 from the biobrick distribution 2013 plate 2, well L2.
- => The fragment was gel extracted with QIAquick gel extraction kit, digested with DpnI, and gel extracted again, resulting in final concentrations of 10.7 ng/µl for FS_77 and 4.8 ng/µL for HM_20
- => PCR will be repeated with several samples from the colonies transformed with pSB6A1 from the biobrick distribution 2013 plate 2, well L2
PCR Conditions BB.W21.A
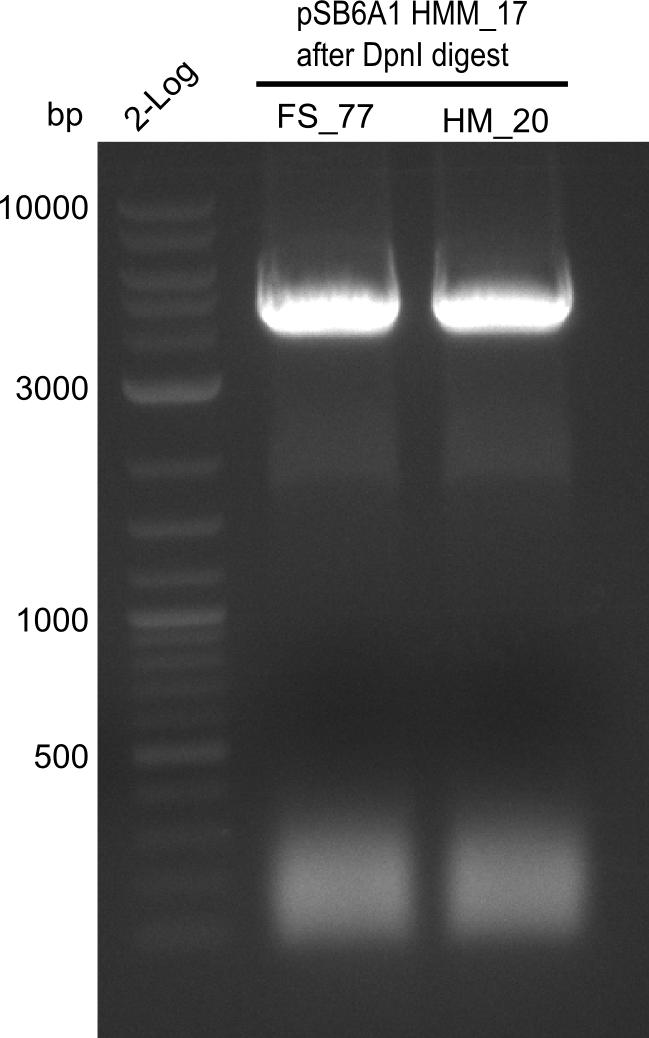
Amplification of backbone fragment pSB6A1 from colonies of DH10ß, transformed with pSB6A1 from the biobrick distribution 2013 plate 2, well L2
4x PCR with HM_20 and 4x PCR with FS_77
| Reagent | BB pSB6A1 | BB pSB6A1 |
|---|---|---|
| Expected length [Kb] | 4.4 | 4.4 |
| Template | colony of DH10B transformed with pSB6A1 plate 5, well K1 | colony of DH10B transformed with pSB6A1 plate 5, well K1 |
| Primer 10 µM fw | 1 µl HM_17 | 1 µl HM_17 |
| Primer 10 µM rev | 1 µl FS_77 | 1 µl FS_77 |
| Phusion Master Mix (2x) | 10 | 10 |
| DMSO | 1 | 1 |
| ddH2O | 6 | 6 |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 80 | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Result
Expected band: 4.4 Kb
Gel shows amplification of fragment.
- => Fragment was cut and gel extracted, then DpnI digested and gel purified again.
Generation of DelH Plasmid pHM04 18-09
Gibson Assembly
| Fragment | Concentration [ng/µl] | Amount with BB16 (FS77) [µl] | Amount with BB16 (HM20) [µl] |
|---|---|---|---|
| DN11-FS66 | 172.5 | 1.77 | 1.84 |
| FS67-FS68 | 120.7 | 2.15 | 2.24 |
| FS69-FS70 | 211.3 | 2.12 | 2.21 |
| FS71-HM08 | 146 | 1.24 | 1.30 |
| BB16 amplified with FS77 | 43.1 | 2.72 | - |
| BB16 amplified with HM20 | 35.8 | - | 2.42 |
- Incubate for 1 h at 50°C
Electroporation
- Afterwards 3 µl of each Gibson assembly were taken and 2 µl ddH2O were added
- With 17 µl of the Gibson assembly, isopropanol purification was performed, DNA was eluted in 20 µl ddH2O
- 6 different electroporations in E.coli BL21 DE3 were performed and grown at RT (see scheme below)
| Electroporation name | Inserted amount of Gibson Assembly | Plated out on agar plates |
|---|---|---|
| HM20 1 | 3 µl (Gibson + ddH2O) | * 10 µl * rest |
| HM20 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
| FS77 1 | 1 µl (Gibson + ddH2O) | * 10 µl * rest |
| FS77 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
Colony-PCR CP.W21.A
| Template | 96x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12H DelH I |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| One-Taq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
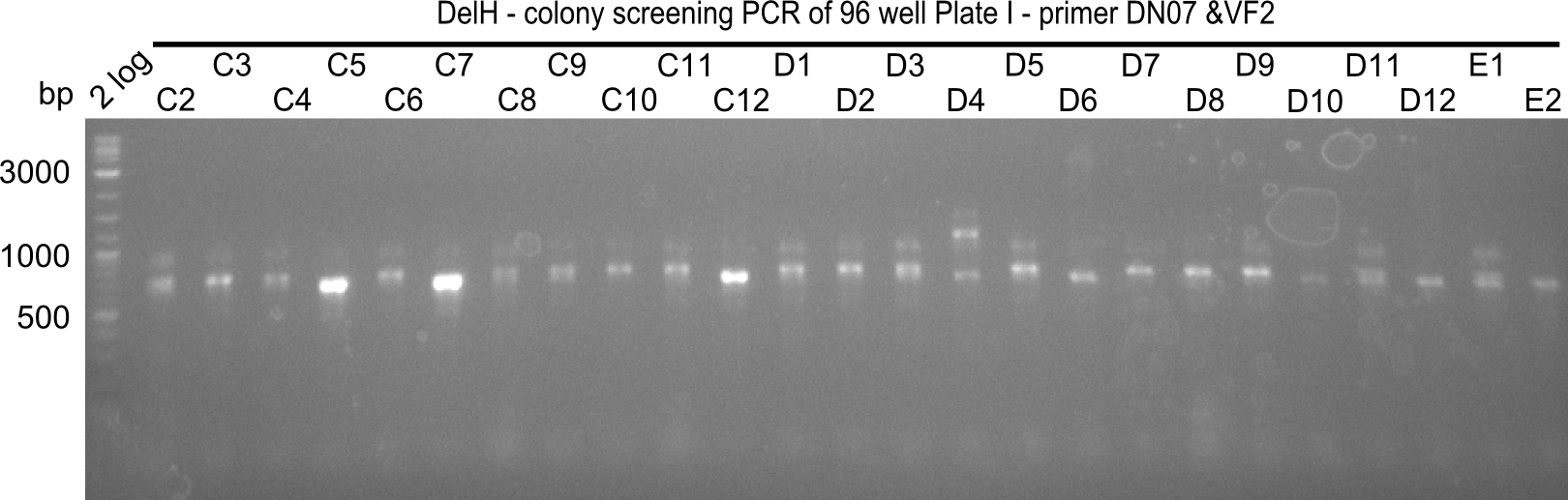
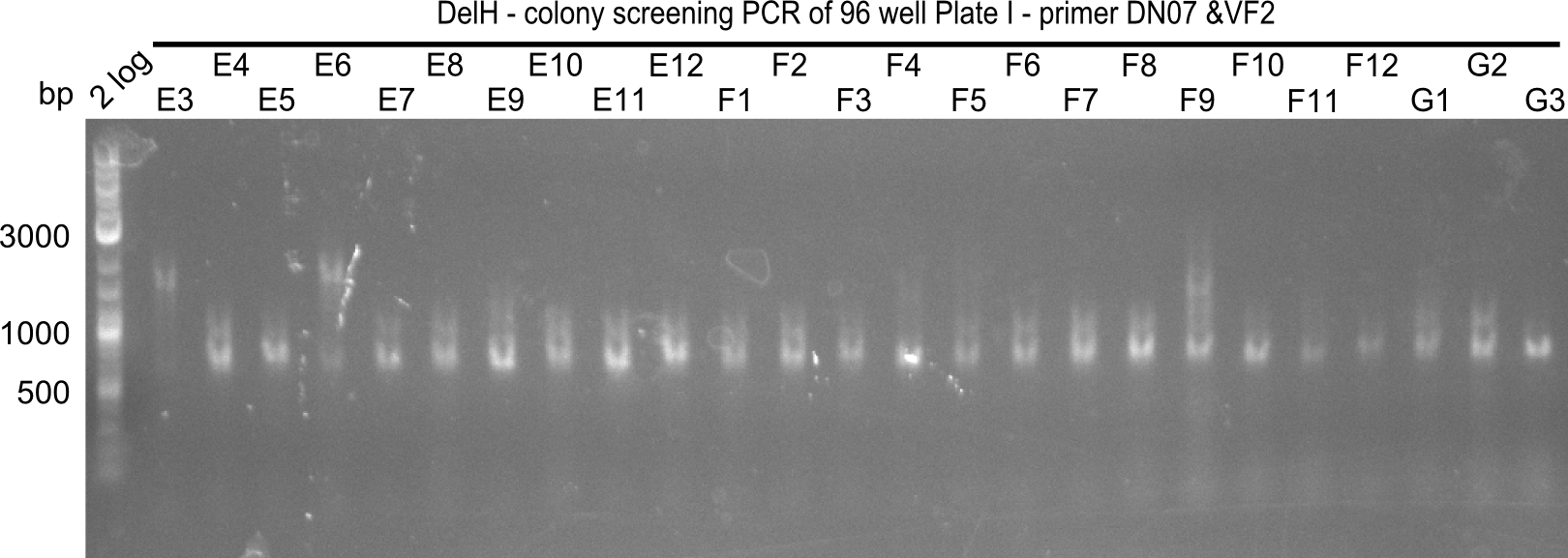
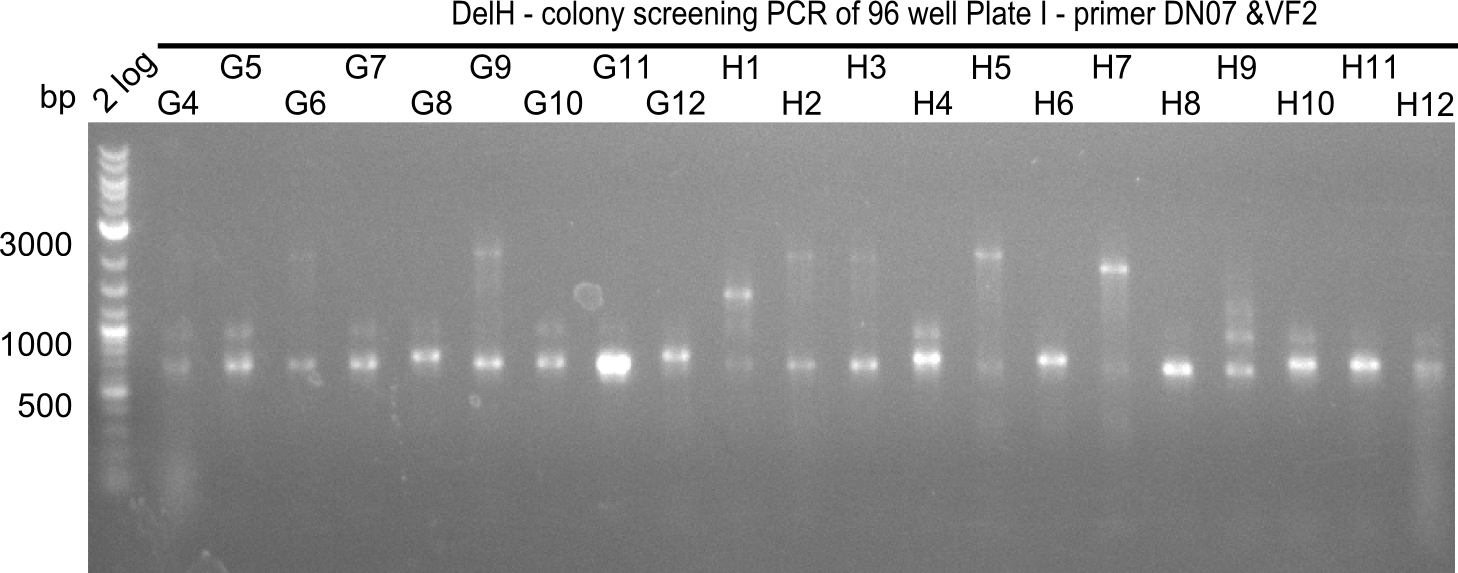
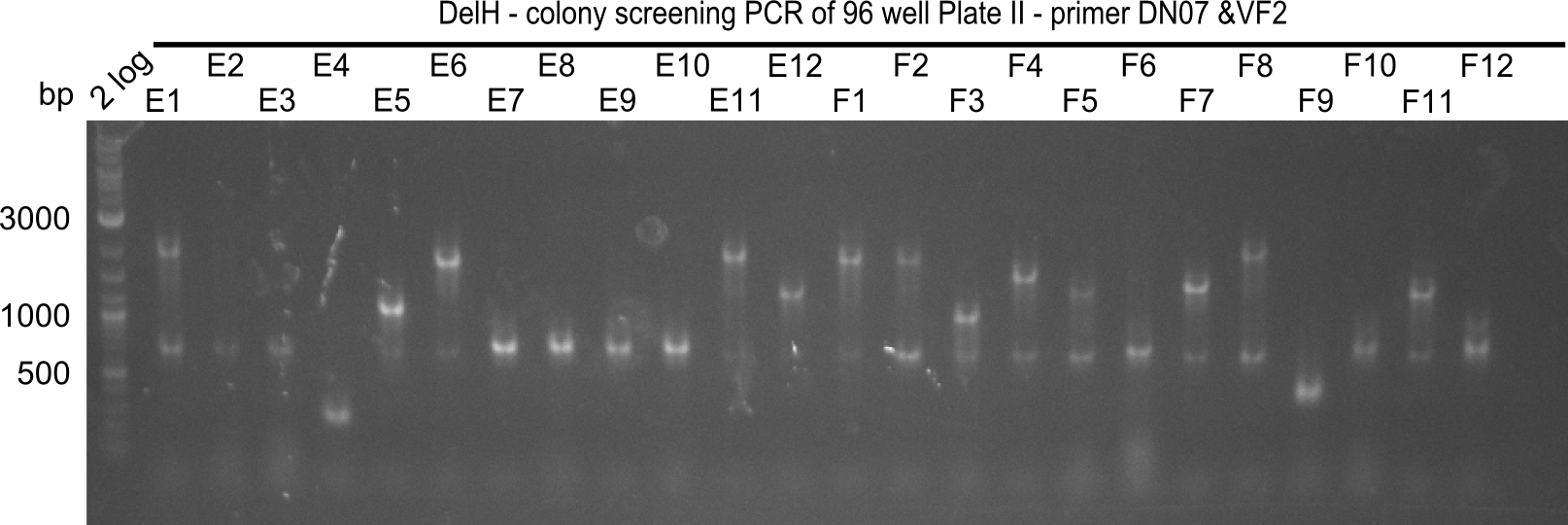
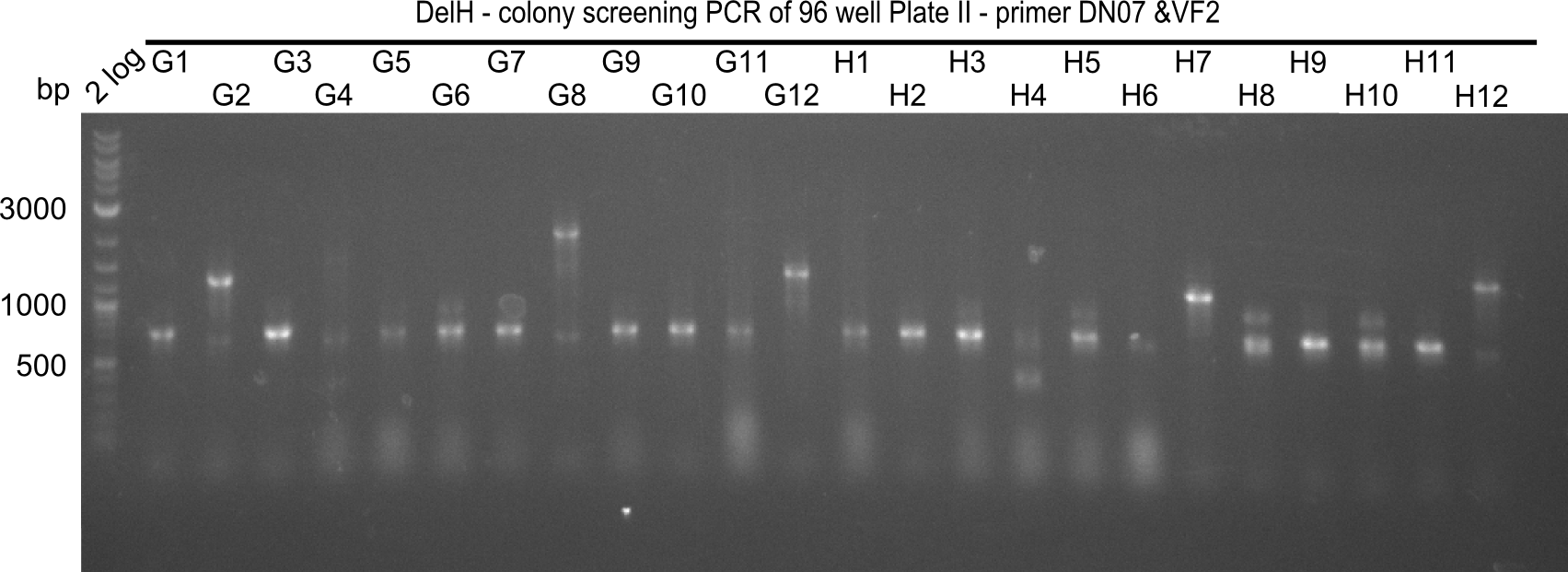
Result
Expected band: 663 bp
The following colonies seem positive, because they show the expected band at 663 bp
| Picture | Positive colonies |
|---|---|
| 21.12 | - |
| 21.13 | C3, C5, C7, C11, C12, D1, D2, D3, D5, D6, D7, D8, D9, D12 |
| 21.14 | E2, E9, F4, F11 |
| 21.15 | G3, G6, G11, G12, H6, H8, H11 |
Colony-PCR CP.W21.B
| Template | 96x 1 picked colony |
| Expected length [bp] | 663 |
| Named | 1A - 12H, DelH II |
| Primer fw 10 µM | 100x 2 µl VF2 |
| Primer rev 10 µM | 100x 2 µl DN07 |
| One-Taq Polymerase (2x) | 100x 10 µl |
| ddH2O | 96x 6 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 95 | 120 |
| 12 | 95 | 30 |
| 68 (touchdown -0.5°C) | 30 | |
| 72 | 45 | |
| 18 | 95 | 30 |
| 65 | 30 | |
| 72 | 45 | |
| 1 | 12 | inf |
Result
Expected band: 663 bp
The following colonies seem positive, because they show the expected band at 663 bp
| Picture | Positive colonies |
|---|---|
| 21.16 | A6, A7, A10, B2, B6 |
| 21.17 | C2, C4, C6, C8, C9, C10, D2, D3, D8, D9 |
| 21.18 | E7, E8, E9, E10, F6 |
| 21.19 | G1, G3, G6, G7, G9, G10, H2, H3, H8, H11 |
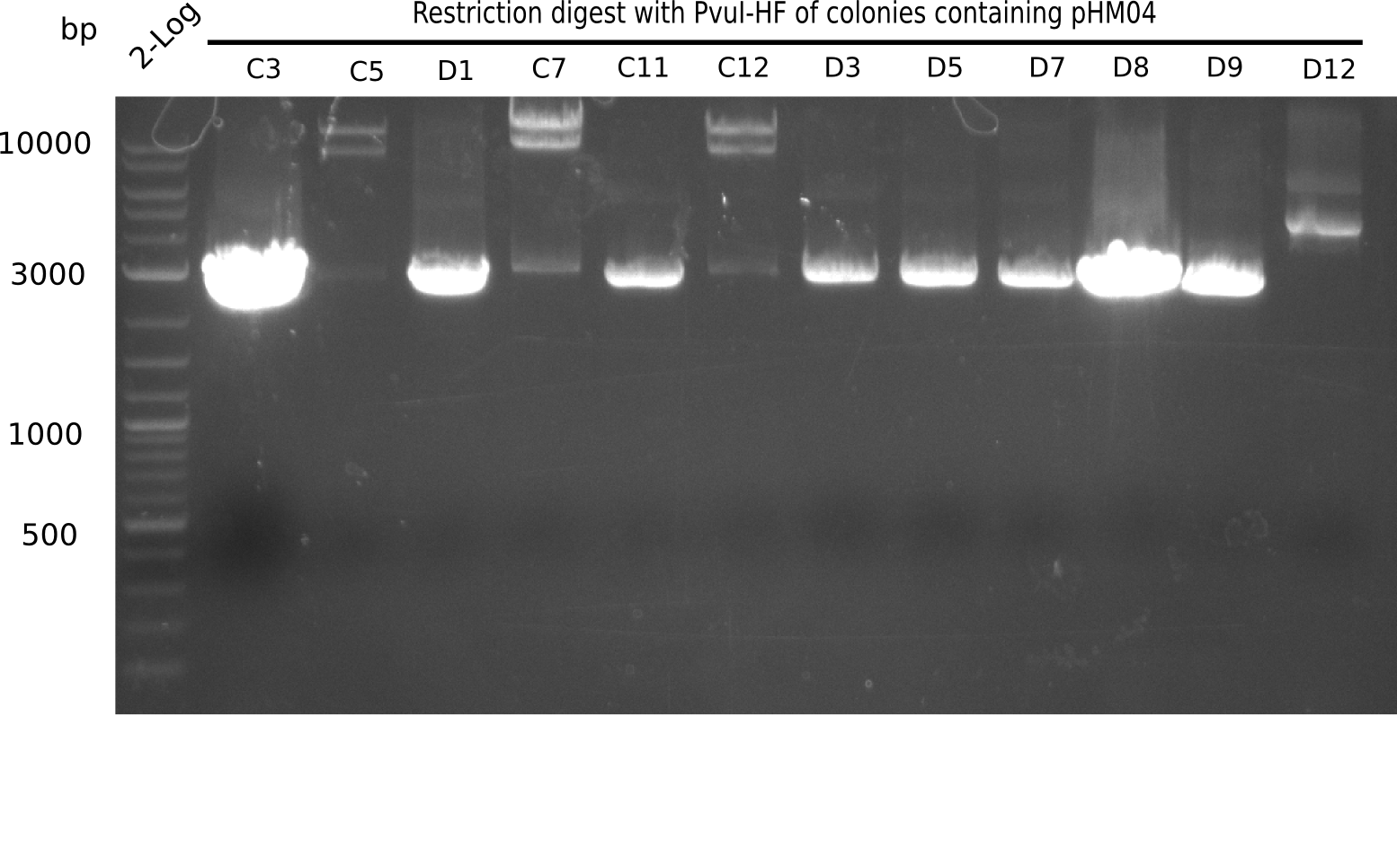
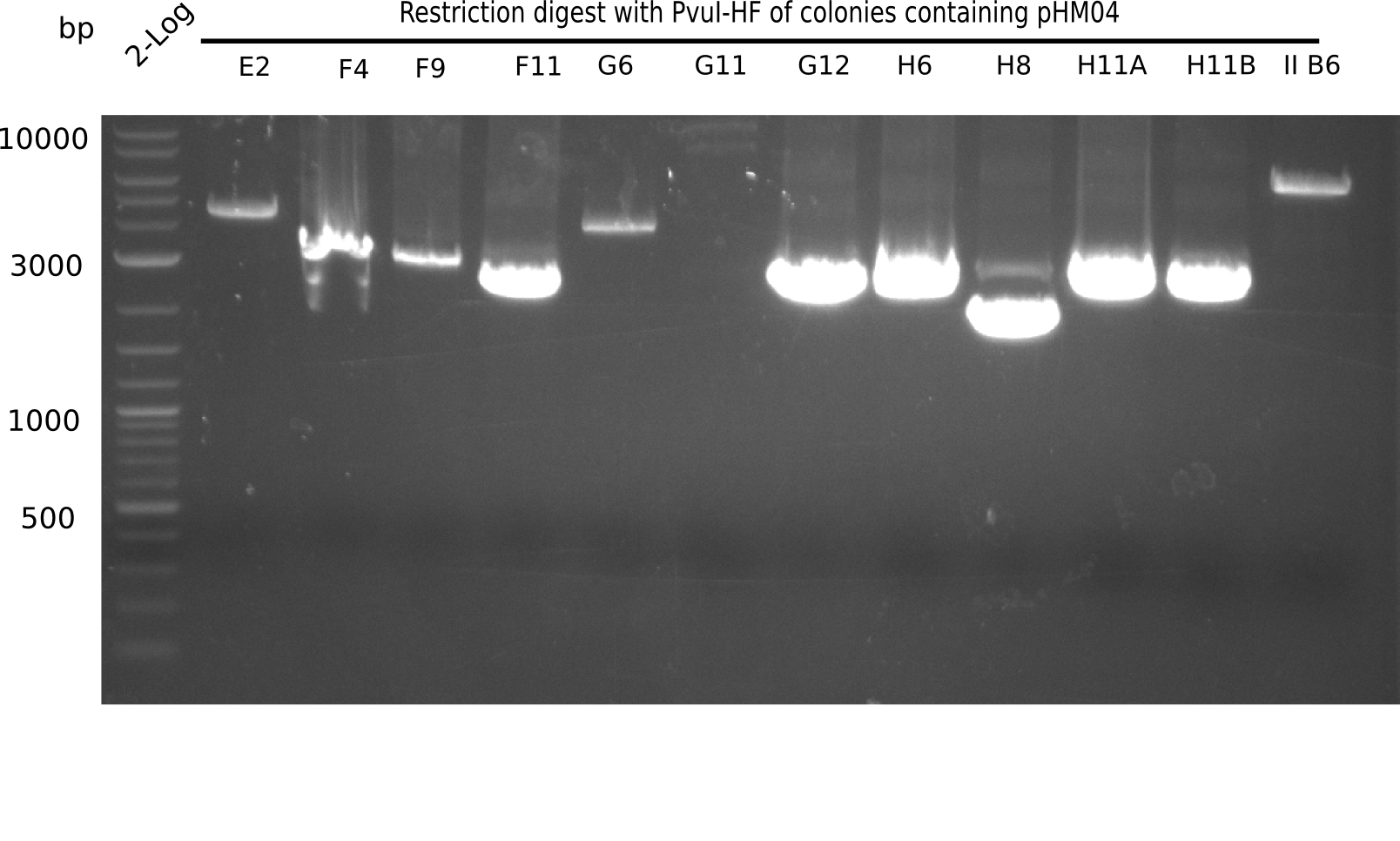
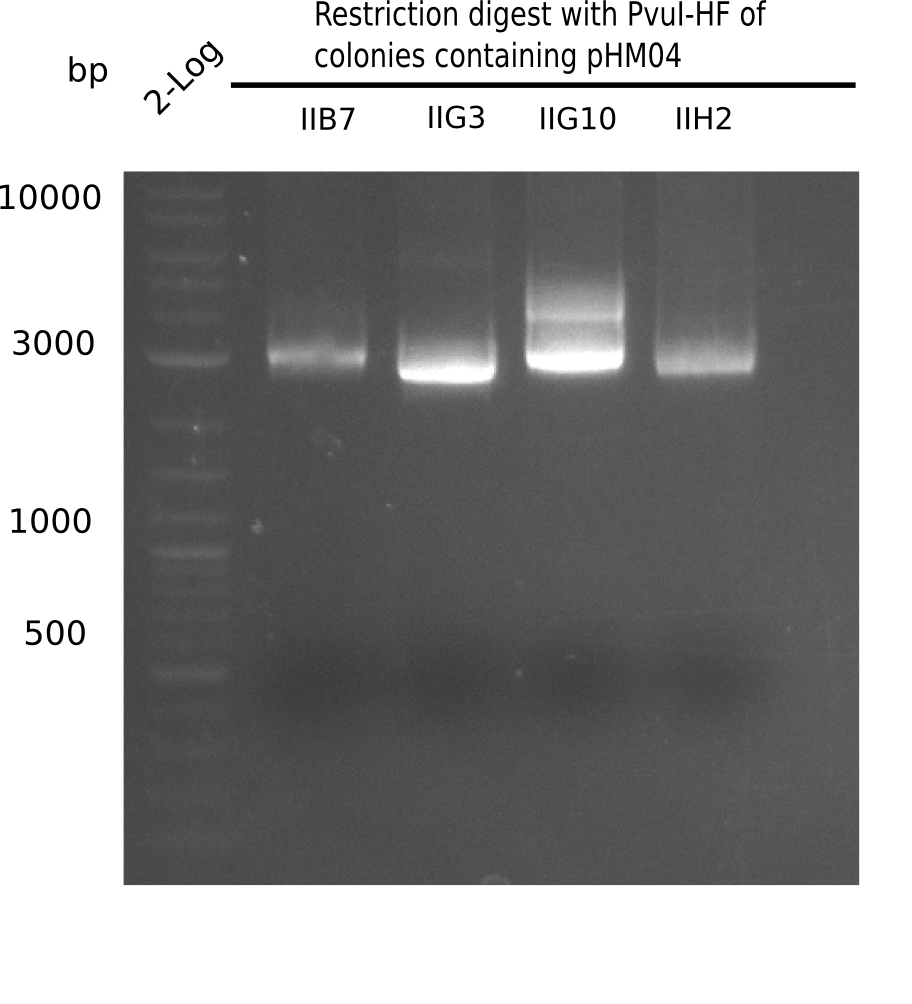
Test Restriction Digest
The positive screened colonies were digested with PvuI.
| Template DNA | CutSmart Buffer | PvuI | ddH2O | Total Volume |
|---|---|---|---|---|
| 8.5 µl 1 µl | 0.5 µl | - | 10 µl |
Result
Expected bands: 11,621, 8,628 & 2,685 bp
The following colonies show the correct pattern:
| Colonies | Positive in Digest | Concentration in Midiprep [ng/µl] |
|---|---|---|
| I C5 | + | 454 |
| I C7 | + | 1312 |
| I C12 | + | 5995 |
| I G11 | + | 3887 |
| II A6 | + | 1017 |
| II D2 | + | 926 |
Sequencing
Clones C5, C7, C12, G11, II A6 and II D2 were send for sequencing with VF2.
Result
| Colonies | Alignment File |
|---|---|
| I C5 | File:Heidelberg Sequencing Result pHM04 DN07 colony 05 old.clustal.txt |
| I C7 | sequencing insufficient |
| I C12 | sequencing insufficient |
| I G11 | discarded |
| II A6 | discarded |
| II D2 | discarded |
Amplification of Backbone
PCR Conditions BB.W21.B
Amplification of backbone parts registry well 2L, transformed 19-09 using HPLC purified HM11
| Reagent | HM11 |
|---|---|
| Template | 1 µl of minipreped pSB6A1 (2L) |
| Primer fw 10 µM | 2 µl HM11 |
| Primer rev 10 µM | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 1 |
| 68 (touchdown -0,5°C) | 5 | |
| 72 | 90 | |
| 18 | 98 | 1 |
| 68 | 5 | |
| 72 | 90 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
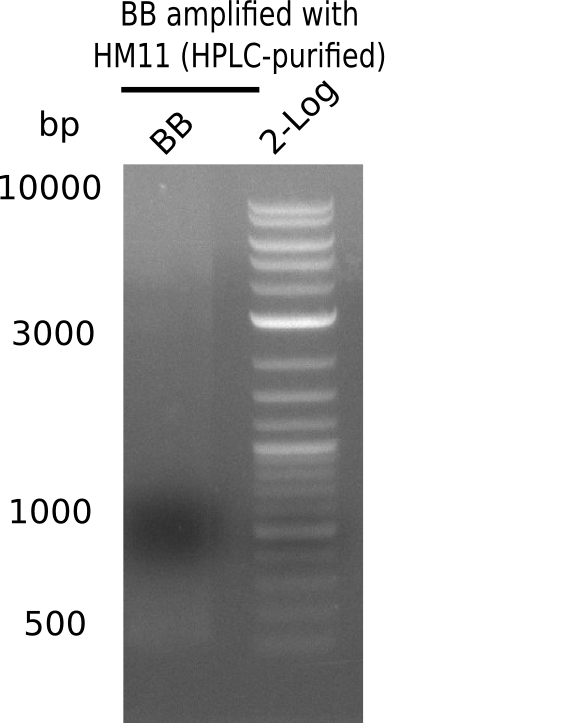
Result
Expected band: 4.4 Kb
Gel does not show amplified backbone fragment.
- => Further optimize PCR conditions.
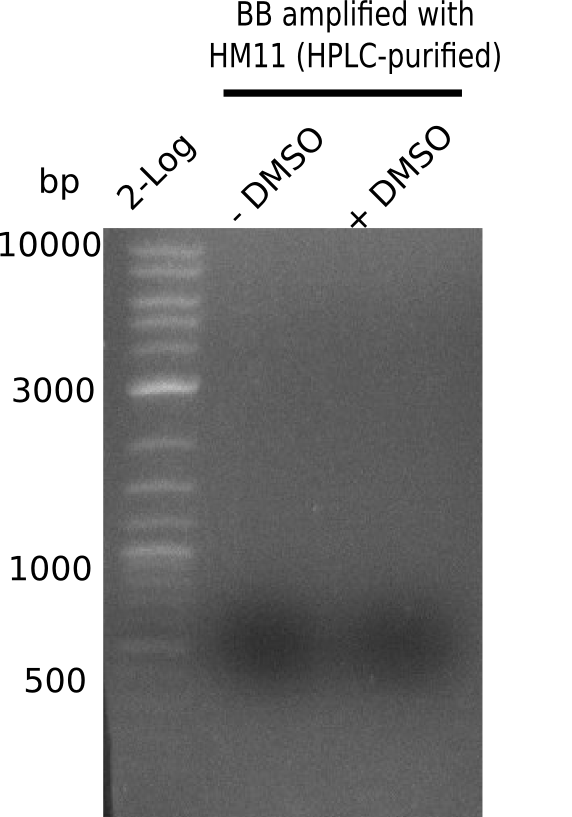
PCR Conditions BB.W21.C
| Reagent | HM11 |
|---|---|
| Template | 1 µl of minipreped pSB6A1 (2L) |
| Primer fw 10 µM | 2 µl HM11 |
| Primer rev 10 µM | 2 µl HM17 |
| Phusion Flash Ready Mix | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 30 | 98 | 1 |
| 68 | 5 | |
| 72 | 90 | |
| 1 | 72 | 5 min |
| 1 | 4 | inf |
Result
Expected band: ~ 4.4 Kb
Gel shows strong and specific amplification of backbone.
- => Fragments were cut and gel isolated.
Generation of DelH Plasmid pHM04 20-09
Gibson Assembly
| Fragment | Concentration [ng/µl] | Amount with BB16 (FS77) [µl] | Amount with BB16 (HM20) [µl] |
|---|---|---|---|
| DN11-FS66 | 172.5 | 1.77 | 1.84 |
| FS67-FS68 | 120.7 | 2.15 | 2.24 |
| FS69-FS70 | 211.3 | 2.12 | 2.21 |
| FS71-HM08 | 146 | 1.24 | 1.30 |
| BB16 amplified with FS77 | 43.1 | 2.72 | - |
| BB16 amplified with HM20 | 35.8 | - | 2.42 |
- Incubate for 1 h at 50°C
Electroporation
- Afterwards 3 µl of each Gibson assembly were taken and 2 µl ddH2O were added
- With 17 µl of the Gibson assembly, isopropanol purification was performed, DNA was eluted in 20 µl ddH2O
- 6 different electroporations in E.coli DH10ß were performed and grown at RT (see scheme below)
| Electroporation name | Inserted amount of Gibson Assembly | Plated out on agar plates |
|---|---|---|
| HM20 1 | 3 µl (Gibson + ddH2O) | * 10 µl * rest |
| HM20 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
| FS77 1 | 1 µl (Gibson + ddH2O) | * 10 µl * rest |
| FS77 20 | 20 µl (isopropanol purified) | * 10 µl * rest |
New HPLC Purified Primers
As pointed out before, we suspect DelH to be toxic for E. coli and therefore, only colinies containing mutated constructs survive. To avoid this, we ordered new shorter and HPLC purified primers for the assembly of pHM04.
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM_20:BB_HPLC_rev | 11-09-2013 | HPLC version of HM11 Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | GATTTGGCGCAGGCGGCCACGGTCCATctagtatttctcctctttc |
| FS_77:BB_HPLC_rev | 11-09-2013 | Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | GCGATTTGGCGCAGGCGGCCACGGTCCATCTAGTATTTCTCCTCTTTC |
Mutagenesis of DelH clones I 6B and 15
Strategy
As our clones which have the DelH construct keep on having mutations and deletions we decided to fix this problem by carrying out site-directed mutagenesis. The targeted mutations are the deletion of parts of the ribsome binding site in clone I6B and the deletion at the beginning of DelH in clone 15. For this we make use of two restriction enzymes (MfeI & XbaI) which both only cut in the construct once. By this we excise the fragment with the mutation. The larger fragment (17.3 kbp) without mutation is kept.
Furthermore we ordered four new primers. The smaller fragment is then PCR amplified with those primers in two fragments. Primer HM22 and HM23 cure the mutations. With those primers also recognitions sites for the restriction enzyme BbsI are introduced, which is needed for religation of the two fragments. BbsI cuts two basepair after its recognition site.
HM21 and HM24 are the corresponding primer. They introduce XbaI and MfeI sites as well as in each on BbsI recognition site. Those are needed for religation.
In the end the two fragments are both digested with BbsI. In the adjacent ligation the two fragments can ligate together due to the BbsI recognition sites. Furthermore BbsI opens the introduced MfeI and XbaI sites through which the two fragments can ligate with the digested 17.3 kb fragment.
However the XbaI site is not introduced correctly again. Therefore we can test if the mutagenesis had been successful by digesting with XbaI. If the site is not present anymore we successfully got rid of the mutations.
New Primers
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| HM21_fw_lacI_BbsI_Xba | 2013-09-15 | Forward primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA CTAGGCAATACGCAA |
| HM22_rev_RBS | 2013-09-15 | Reverse Primer in RBS for mutagenesis | TTTTGAAGACAA CTCTTTCTCTAGTATGTGTGAAATTG |
| HM23_fw_RBS | 2013-09-15 | Forward Primer in RBS for mutagenesis | TTTTGAAGACAA AGAGGAGAAATACTAGATGGACCGTGGC |
| HM24_rev_BbsI_MfeI | 2013-09-15 | Reverse primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA AATTGGACAGCGCGGCATGCCGGTTG |
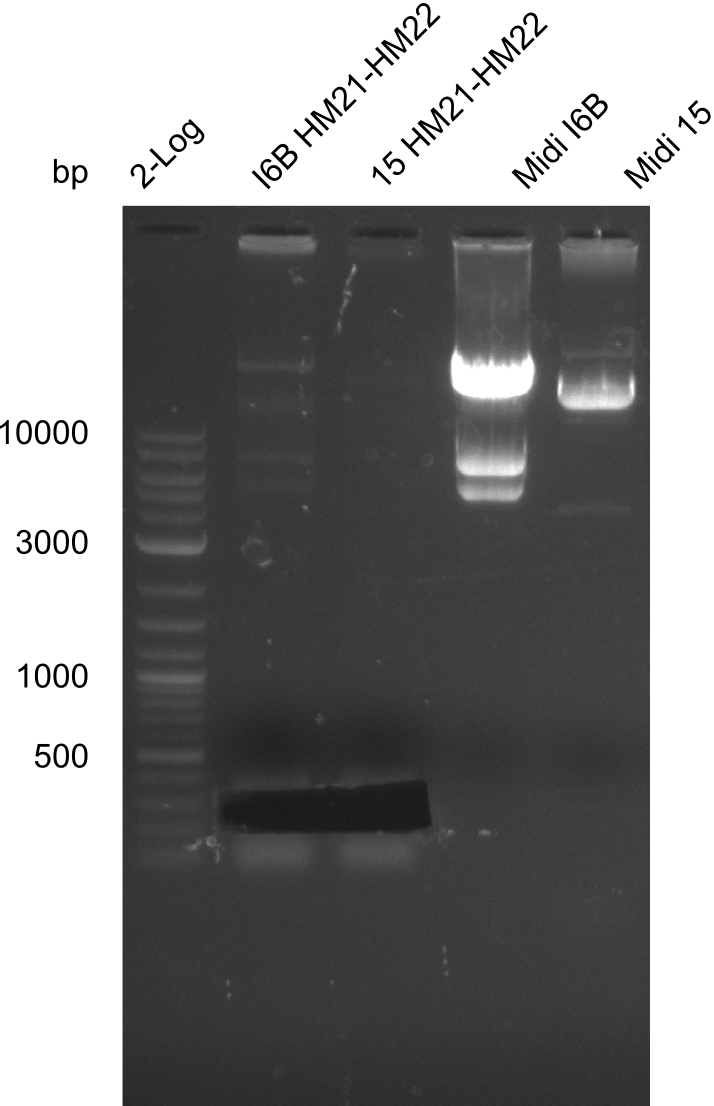
Purification of Midiprep of Clone I6B
As can be seen in figure 21.23, the midi-prep of colony I 6B is contaminated. Thus, we tried to purify only the largest band by gel extraction with the QIAEX II Gel Extraction Kit (Qiagen). The procedure was as following:
- Application of 2 µl of midiprep on gel (4596.7 ng/µl --> ~10 µg)
- Excision of band at about 17.4 kb
- Gel extraction of DNA with QIAEX II Gel Extraction Kit (Qiagen)
Result
- The concentration of the purified fragment was only 4.7 ng/µl after extraction.
- => We are trying to elute again.
- The second elution did not work either
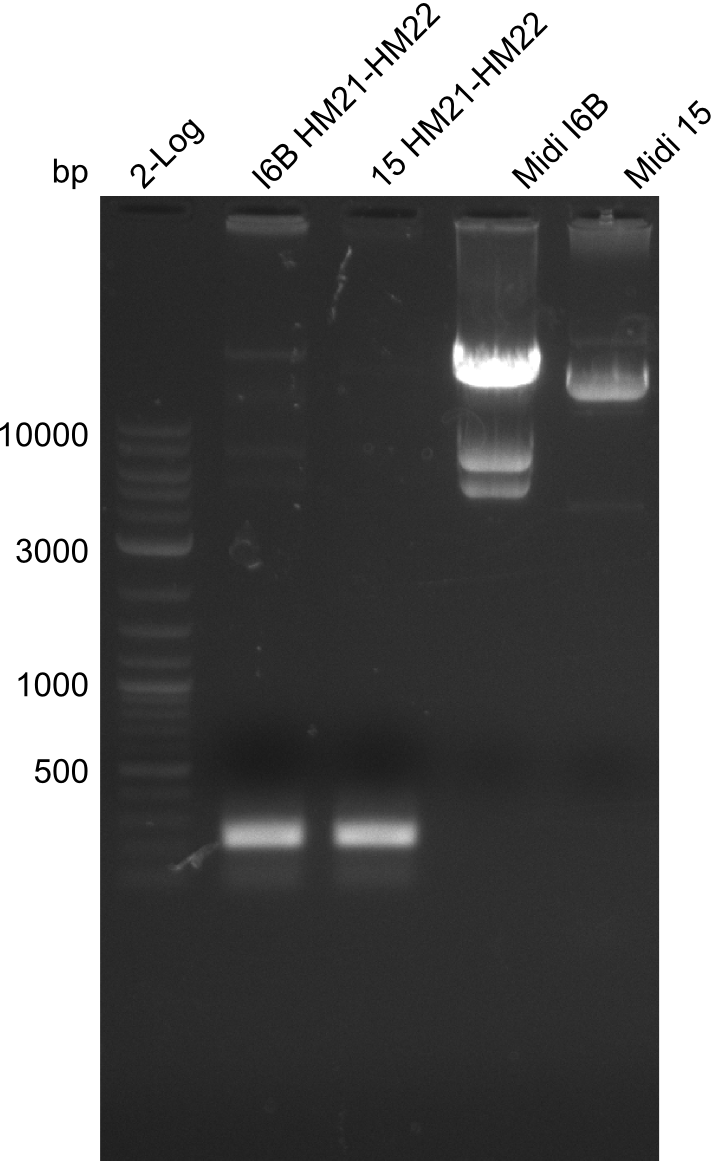
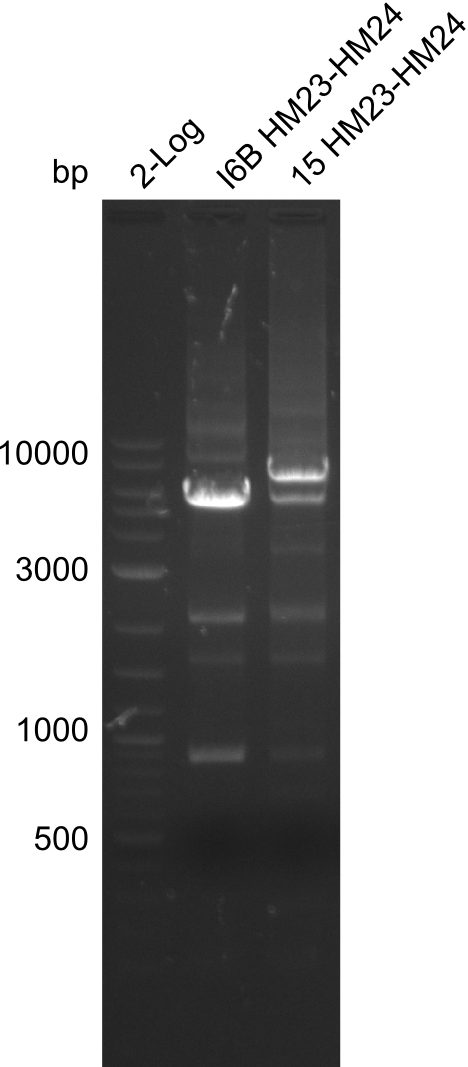
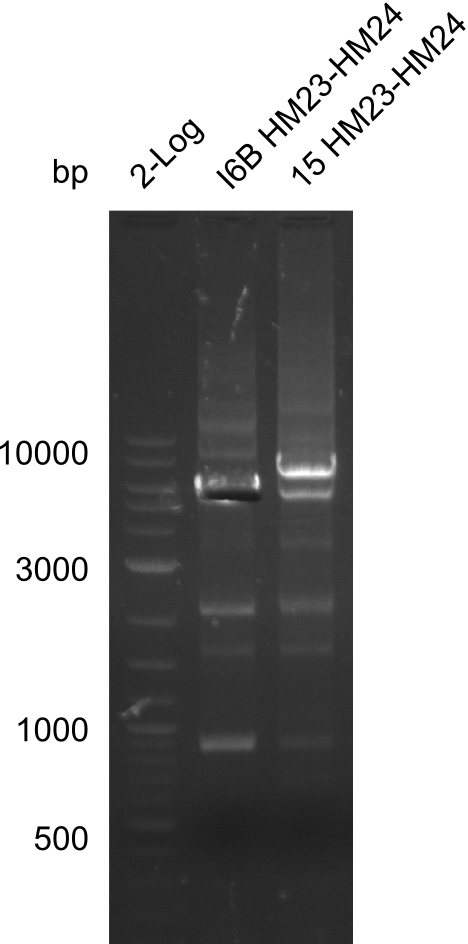
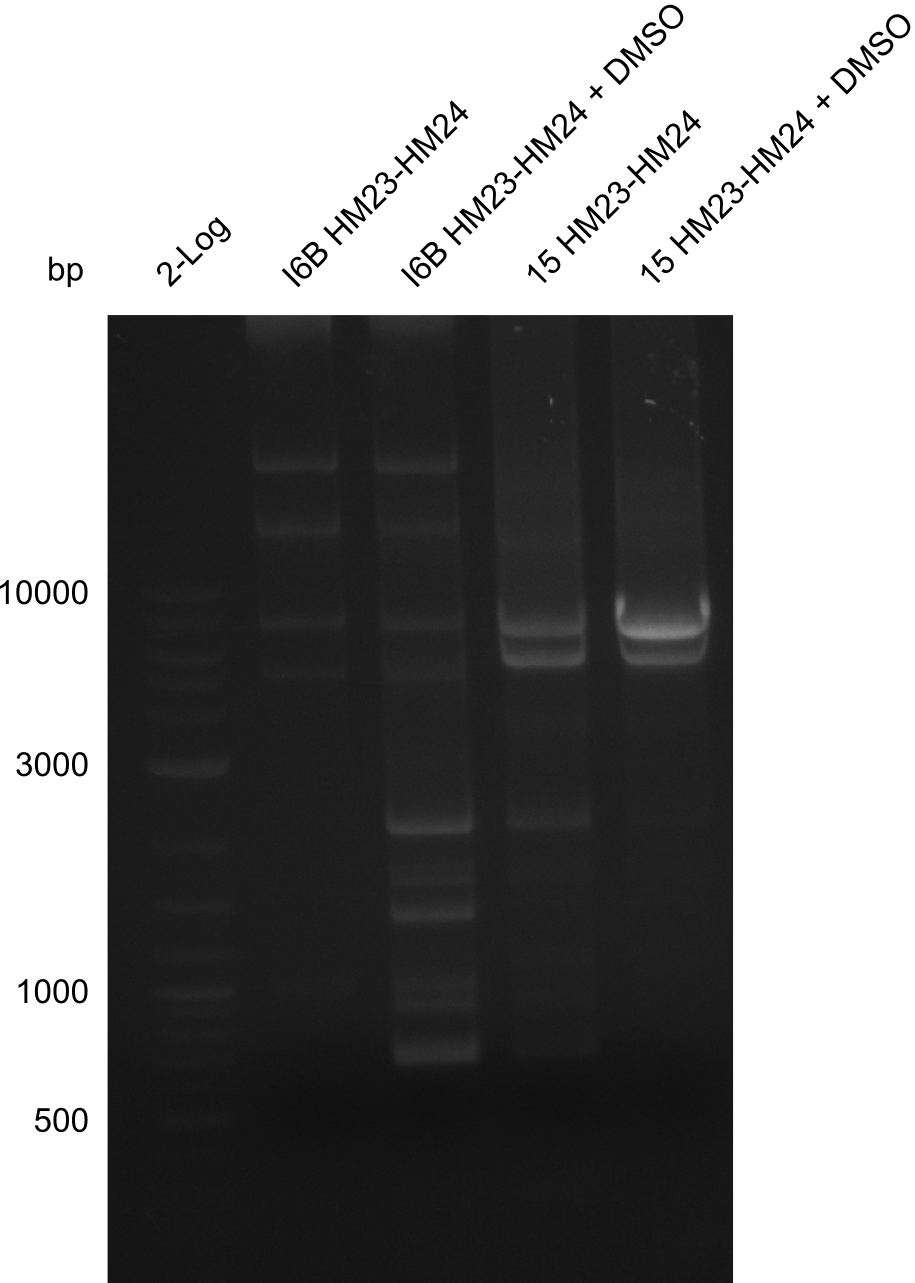
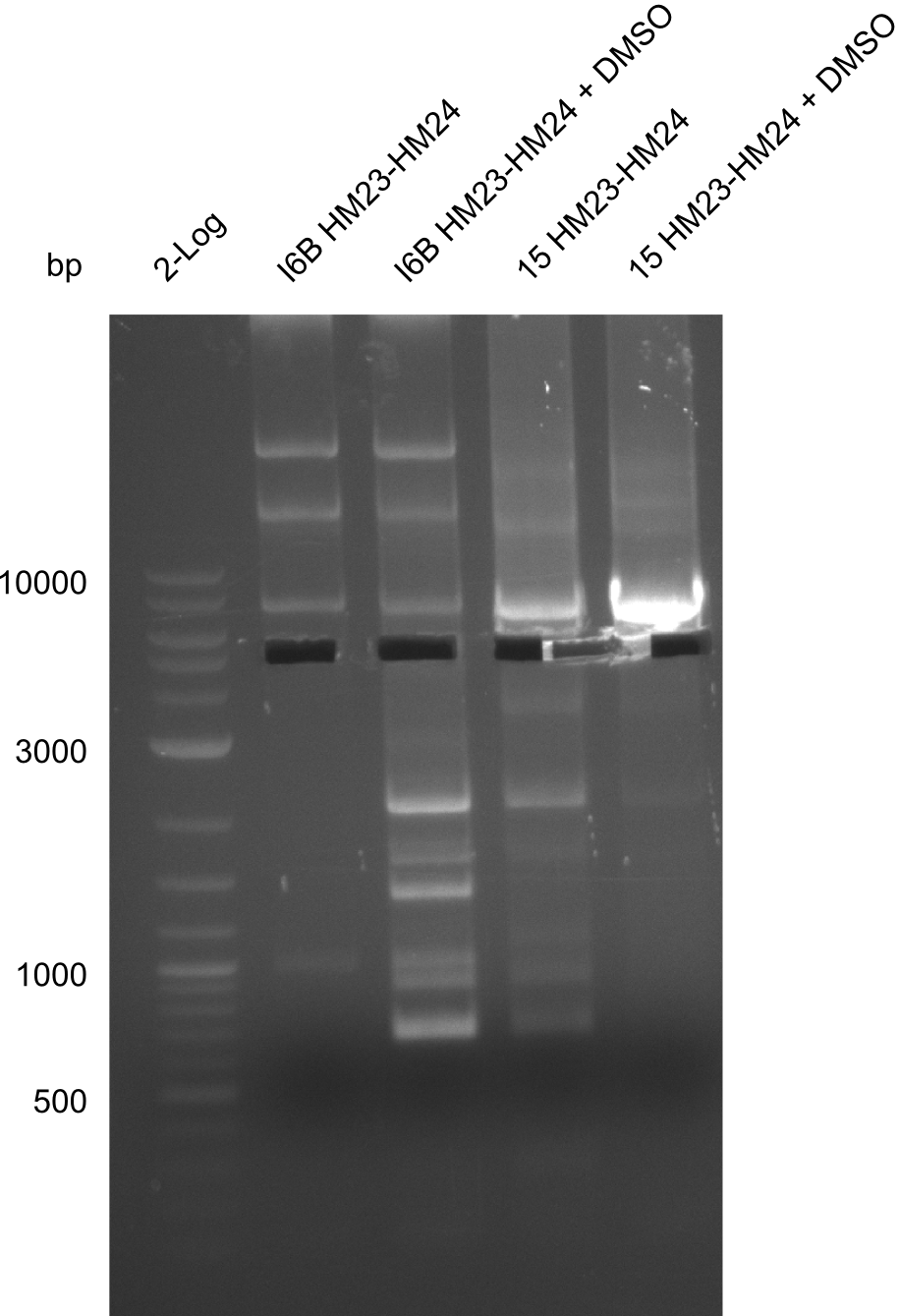
Amplification of HM23 to HM24
1x 20 µl for each colony
| Template | 0.2 µl of either midiprep colony I6B or 15 |
|---|---|
| Expected length [kbp] | 5.3 |
| Named | I6B or 15 HM23-HM24 |
| Primer fw 10µM | 2 µl HM23 |
| Primer rev 10 µM | 2 µl HM24 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 s |
| 12 | 98 | 1 |
| 68 (touchdown -0.5°C) | 5 | |
| 72 | 1:50 min | |
| 18 | 98 | 1 |
| 65 | 5 | |
| 72 | 1:50 min | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 69 | 5 | |
| 72 | 1:50 min | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
| Template | 0.2 µl of either midiprep colony I6B or 15 |
|---|---|
| Expected length [kbp] | 5.3 |
| Named | I6B or 15 HM23-HM24 |
| Primer fw 10µM | 2 µl HM23 |
| Primer rev 10 µM | 2 µl HM24 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 4 µl |
| DMSO | 1 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 s |
| 30 | 98 | 1 s |
| 72 | 1:40 min | |
| 1 | 72 | 7 min |
| 1 | 12 | inf |
Result
Gels for amplification of HM23 to HM24
Amplification of HM21 to HM22
| Template | 0.2 µl of either midiprep colony I6B or 15 |
|---|---|
| Expected length [kbp] | 200 bp |
| Named | I6B or 15 HM21-HM22 |
| Primer fw 10µM | 2 µl HM21 |
| Primer rev 10µM | 2 µl HM22 |
| Phusion Flash (2x) | 10 µl |
| ddH2O | 5 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 63 | 5 | |
| 72 | 10 | |
| 1 | 72 | 5 min |
| 1 | 12 | inf |
Result
Gels for amplification of HM21 to HM22
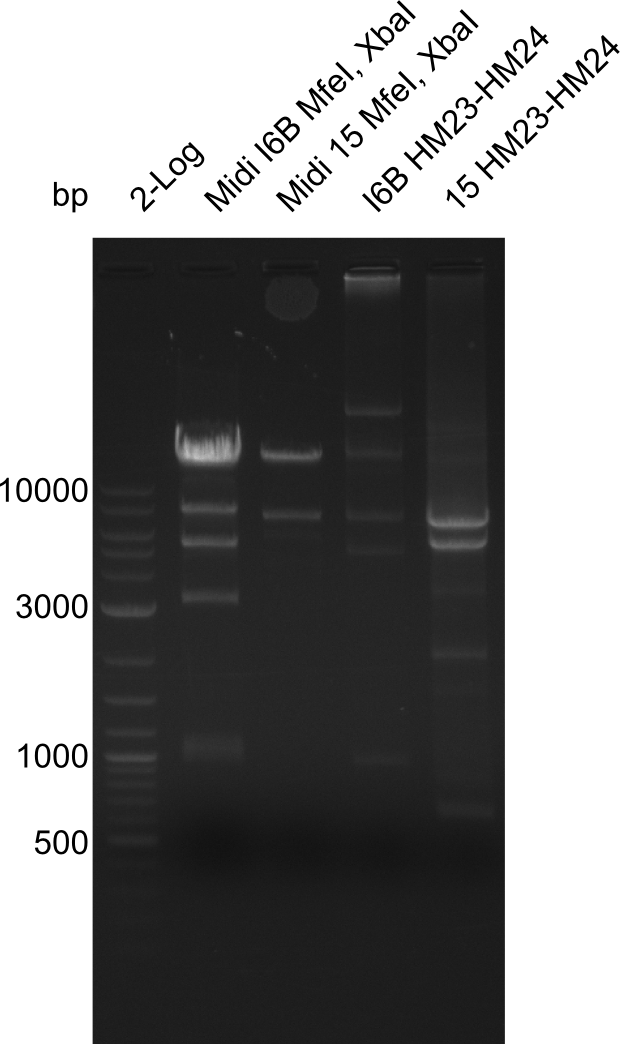
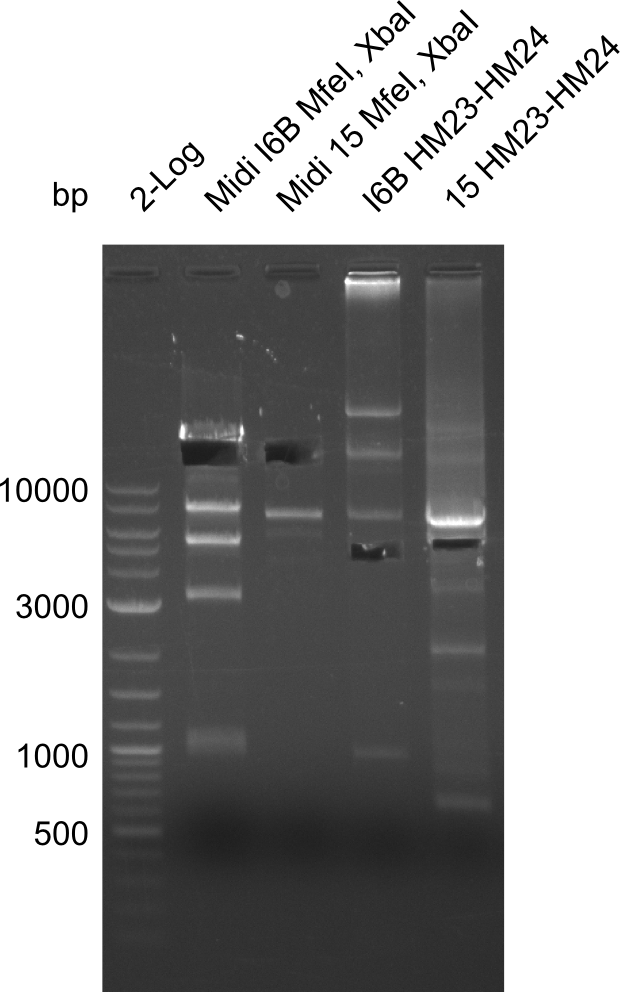
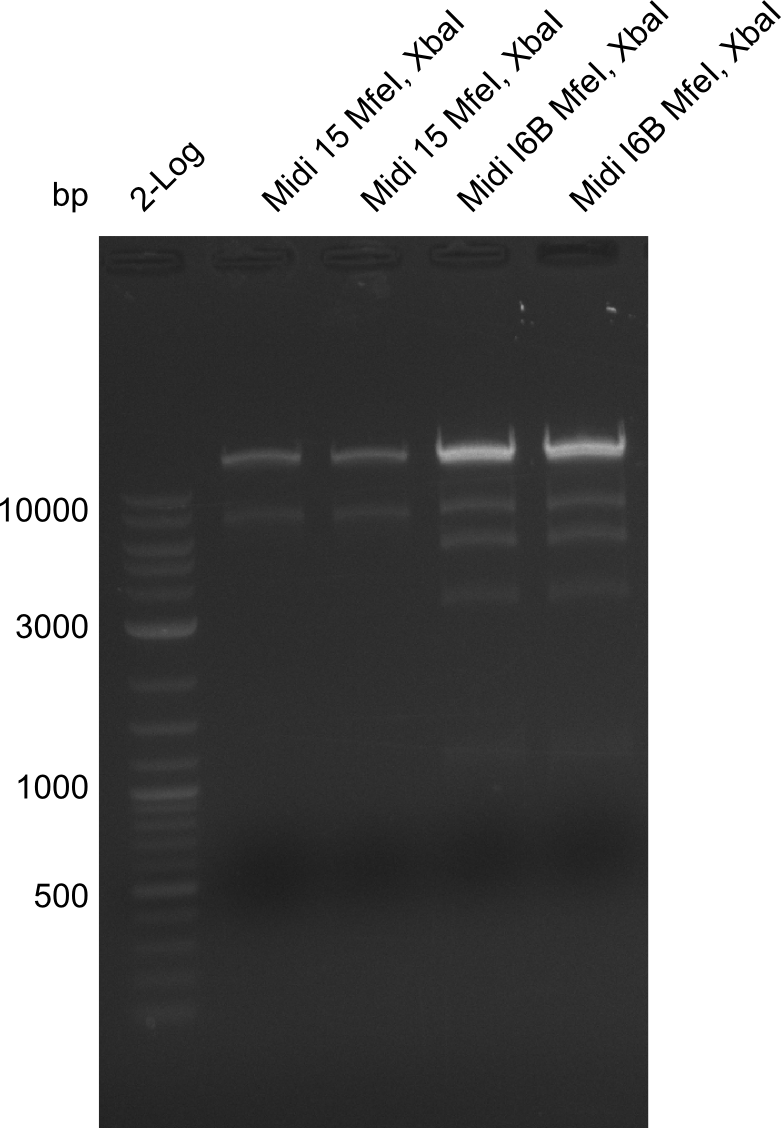
Restriction Digest of pHM04 (I6B) with XbaI and MfeI
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B or 15 | 0.5 |
| Cutsmart buffer | 2 |
| XbaI | 1.5 |
| MfeI-HF | 1.5 |
| ddH2O | 14.5 |
- Incubation for 2 h at 37°C
Result
- Expected band pattern: 17.3 kbp, 5.3 kbp
Gels for restriction Digest of pHM04 (I6B) with XbaI and MfeI
- The MfeI already expired in 2011
- => We will test digest the construct again with a new enzyme
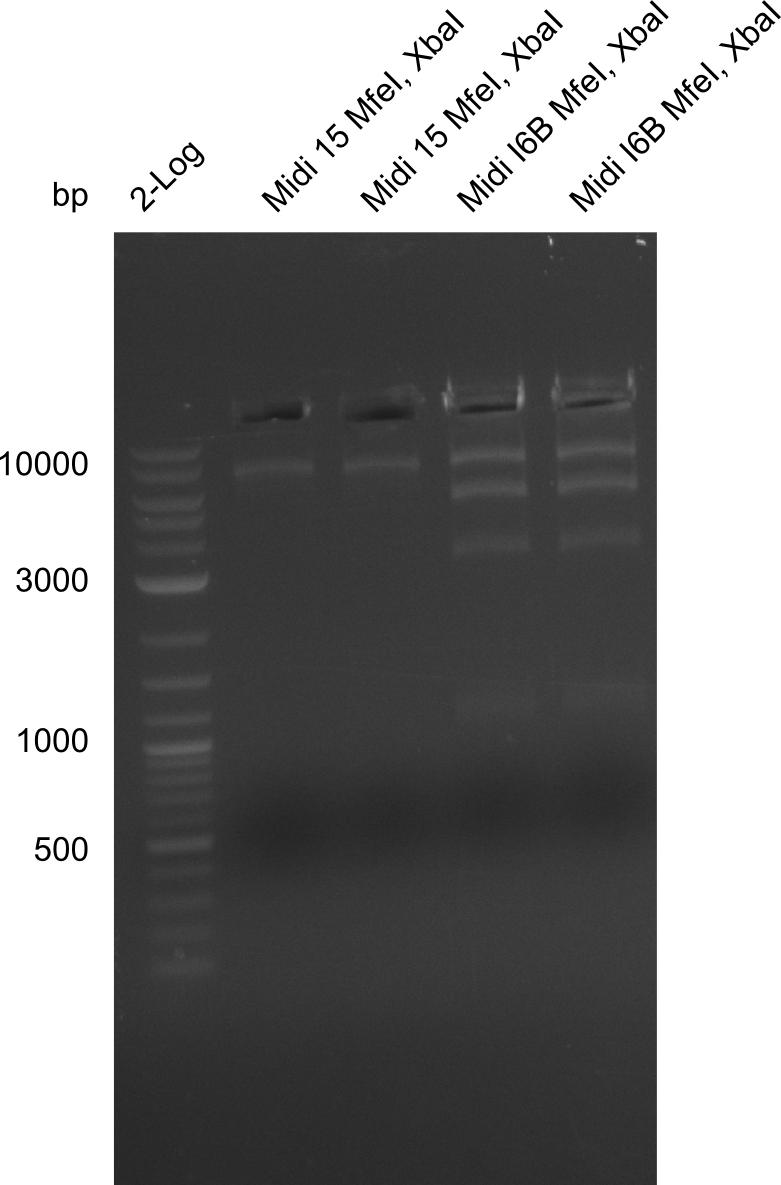
Restriction Digest of pHM04 (I6B) with XbaI and MfeI
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B or 15 | 0.5 |
| Cutsmart buffer | 5 |
| XbaI | 1.5 |
| MfeI-HF | 1.5 |
| ddH2O | 14.5 |
| Incubation for 2 h at 37°C | Expected band pattern: 17.3 kbp, 5.3 kbp |
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B purified 1:10 | 1 |
| Cutsmart buffer | 2 |
| XbaI | 1 |
| MfeI-HF | 1 |
| ddH2O | 15 |
| Incubation at room temperature, over night | Expected band pattern: 17.3 kbp, 5.3 kbp |
Result
Due to the contamination in DelH it was not possible to decide whether the fragment really had been digested. Thus we have to test whether MfeI Cutting site is really present.
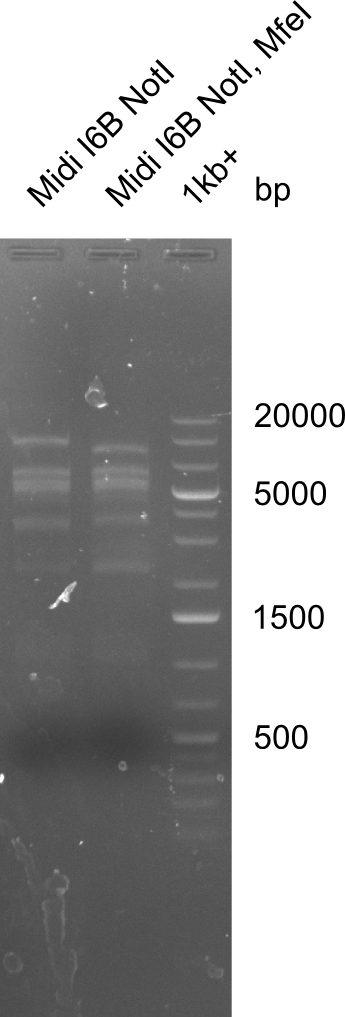
Test for MfeI Cutting Site
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B | 0.5 |
| Cutsmart buffer | 2 |
| NotI-HF | 1 |
| ddH2O | 16.5 |
| Reagent | Volume [µl] |
|---|---|
| Midiprep colony I6B | 0.5 |
| Cutsmart buffer | 2 |
| NotI-HF | 1 |
| MfeI-HF | 1 |
| ddH2O | 15.5 |
Incubation at room temperature over night.
--> As the upper band is lower for the digest with both enzymes the MfeI restriction site is apparently present. Therefore the mutagenesis is tried.
Restriction Digest of Fragment HM21-HM22 (I6B) and HM23-HM24 (I6B) with BbsI
PCR fragments: HM21-HM22 (36.6 ng/µl) and HM23-HM24 (32.4 ng/µl)
| Reagent | Volume [µl] |
|---|---|
| PCR fragment | 20 |
| 2.1 buffer | 5 |
| BbsI | 1 |
| ddH2O | 24 |
Ligation of Digested pHM04 (I6B), HM21-HM22 and HM23-HM24
| Reagent | Volume [µl] |
|---|---|
| digested and purified pHM04 (I6B) | 3 |
| digested HM21-HM22 | 0.5 |
| digested HM23-HM24 | 0.5 |
| T4 ligase buffer | 2 |
| T4 ligase | 1 |
| ddH2O | 13 |
Transformation of Ligation
1 µl and 5 µl were transformed in 50 µl electrocompetent DH10β each by electroporation. The cells were recovered in 400 µl SOC media and plated on LB Amp.
 "
"