Team:Heidelberg/Templates/DelH week4
From 2013.igem.org
Contents |
20-05 - 26-05-13
DelH Fragment F1a
- The new primer arrived.
PCR Conditions F1a.W4.A
| Reagent | DelH F1a | DelH F1a |
|---|---|---|
| Expected length [Kb] | 5 | 5 |
| Template | 1 µl D. acidovorans | 1 µl D. acidovorans |
| Primer fw 10 µM | DN01 | DN01 |
| Primer rev 10 µM | DN08 | DN08 |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | 2.5 µl | - |
| ddH2O | 20,5 | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 s |
| 62 | 5 s | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Results
Expected band: 5 Kb
The gel shows a lot unspecific bands for both DelH F1a samples.
- => Alter PCR conditions.
DelH F1b
- The new primer arrived
PCR Conditions F1b.W4.A
| Reagent | DelH F1b |
|---|---|
| Expected length [Kb] | 5 |
| Template | 1 µl D. acidovorans |
| Primer fw 10 µM | DN07 |
| Primer rev 10 µM | DN02 |
| Phusion Master Mix (2x) | 25 µl |
| DMSO | - |
| ddH2O | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 s |
| 62 | 5 s | |
| 72 | 2:00 min | |
| 1 | 72 | 2:30 min |
| 1 | 4 | inf |
Result
Expected band: 5 Kb
The gel shows a lot unspecific bands for DelH F1b, but also the band at 5 Kb.
- => Band was cut and gel isolated.
Generation of Backbone pSB6A1-AraC-lacZ
Miniprep
- From three different clones, 2ml of pSB6A1-AraC-lacZ (6) in LB Amp were minipreped and eluted in 30 µl ddH2O. Additionally, a glycerol stock was prepared.
Result
| Sample | Concentration [ng/µl] |
|---|---|
| 8 - pSB6A1-AraC-lacZ (6) | 63 |
| 9 - pSB6A1-AraC-lacZ (6) | 61 |
| 10 - pSB6A1-AraC-lacZ (6) | 62 |
PCR Conditions BB.W4.A
| Reagent | 8 - pSB6A1-AraC-lacZ (6) | 9 - pSB6A1-AraC-lacZ (6) |
|---|---|---|
| Expected length [Kb] | 7.382 | 7.382 |
| Template | 1 µl of 1:10 diluted 8 | 1 µl of 1:10 diluted 9 |
| Primer 10 µM fw | 0.5 µl | |
| Primer 10 µM rev | 0.5 µl | |
| Phusion Master Mix (2x) | 25 µl | 25 µl |
| DMSO | - | - |
| ddH2O | 23 µl | 23 µl |
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 16 | 98 | 1 s |
| 66↓ | 5 s | |
| 72 | 3:00 min | |
| 14 | 98 | 1 s |
| 63 | 5 s | |
| 72 | 3:00 min | |
| 1 | 72 | 5:00 min |
| 1 | 4 | inf |
Results
Expected band: 7.4 Kb
The gel shows no bands.
- => Perform test restriction to test identity of plasmid.
Restriction Digest A
- Digest with PstI & XbaI of samples 8, 9 and 10 for checking identity of the construct
| Reagent | Amount [µl] |
|---|---|
| DNA | 10 |
| Enzymes | 1.5 each |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 27 |
- Incubated for 1 h at 37°C
Result
Expected bands: 3360 and 4022 bp
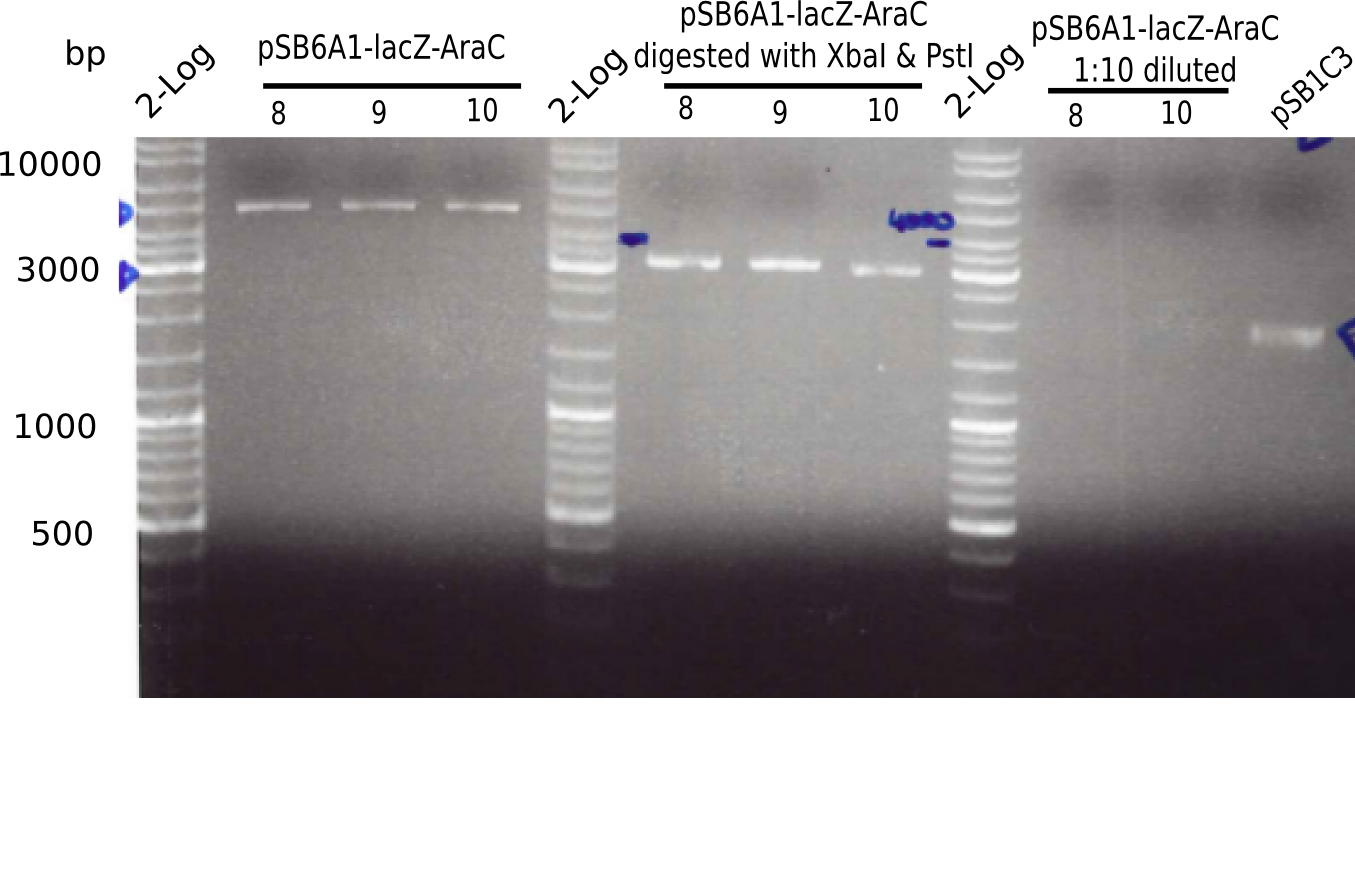
l1:2log ladder, l2: pSB6A1-AraC-lacZ (8), l3: pSB6A1-AraC-lacZ (9), l4: pSB6A1-AraC-lacZ (10),l5:2log ladder, l6: pSB6A1-AraC-lacZ (8) digested with XbaI & PstI, l7: pSB6A1-AraC-lacZ (9) digested with XbaI & PstI, l8: pSB6A1-AraC-lacZ (10)digested with XbaI & PstI, l9:2log ladder, l10: pSB6A1-AraC-lacZ (8) 1:10 diluted, l11: pSB6A1-AraC-lacZ (10) 1:10 diluted, l12: pSB1C3
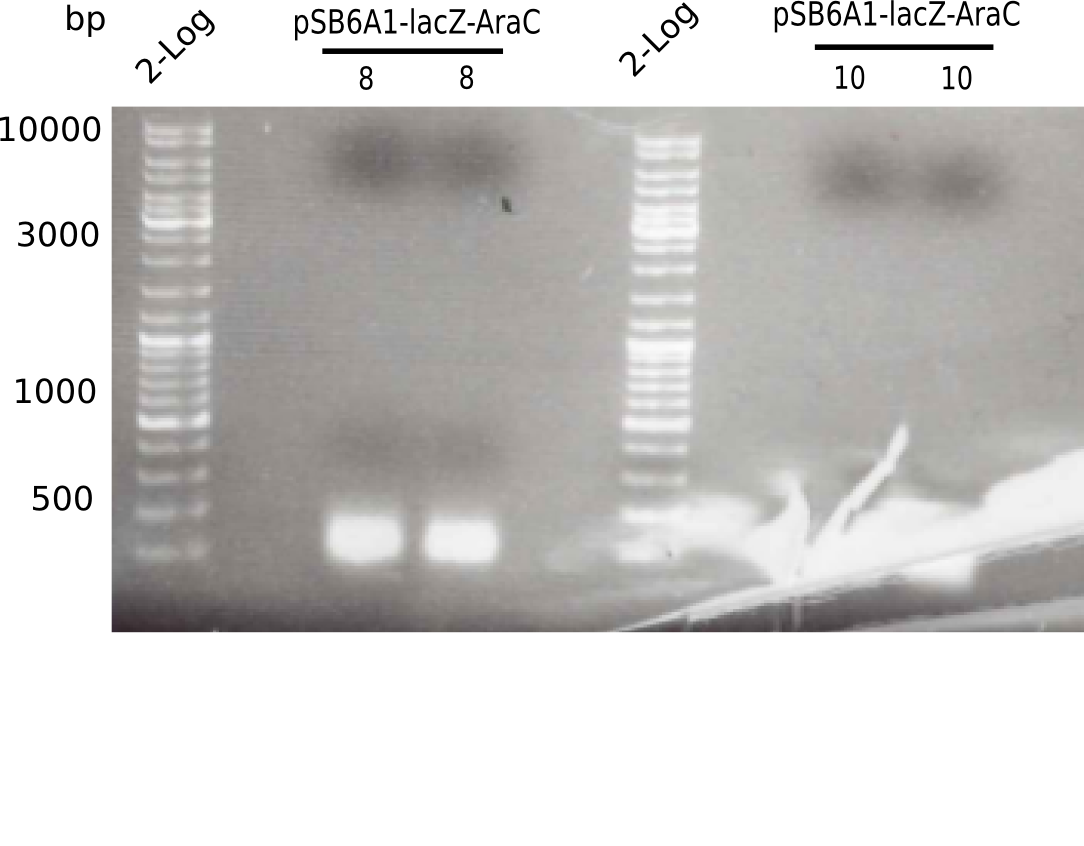
The gel shows a band at 5 Kb.
- => Colonies 8, 9 and 10 are not the desired construct.
Restriction Digest B
- Digest with KpnI of samples 8, 9, 10 for checking length of the construct and KpnI and BamHI to check for identity.
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-AraC-lacZ (8, 9, 10) | 5 |
| Enzymes | 1.5 KpnI |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 33,5 |
| Expected bands | 7.381 bp |
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-AraC-lacZ (8, 9, 10) | 5 |
| Enzymes | 1.5 KpnI & 1,5 BamHI |
| Buffer NEB 3(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 32 |
| Expected bands | 4.607 bp & 2.781 bp |
Result
- Expected bands: 7.381 bp and 4.607 bp & 2.781 bp
- => Colonies 8, 9 and 10 are not desired construct. Pick new colony from pSB1C3-AraC(6)-lacZ and test digest.
Miniprep
A new colony was picked from pSB1C3-AraC(6)-lacZ plate and a mini prep was performed.
Result
pSB6A1-AraC(6)-lacZ construct : 94 [ng/µl]
Test Restriction Digest
The new miniprep as well as one of the wrong pSB6A1-AraC(6)-lacZ ones were restriction digested with EcoRI-HF and PstI.
| Component | Amount [µl] |
|---|---|
| DNA | 5 |
| BSA (10x) | 5 |
| NEB2 buffer (10x) | 5 |
| Enzymes (EcoRI-HF and PstI) | 1.5 each |
| ddH2O | 32 |
- Incubated 1 h at 37°C
Result
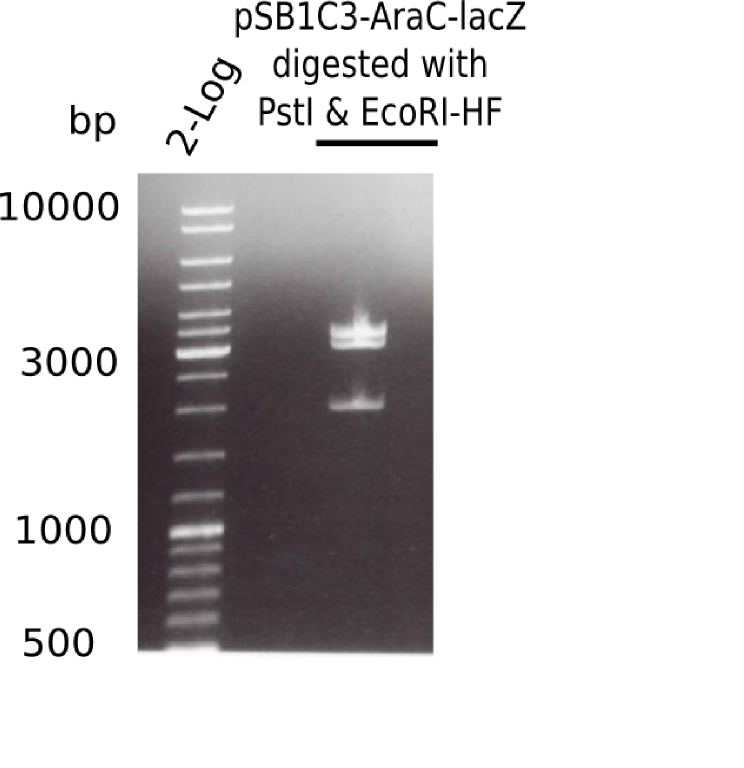
Expected bands: ? Kb
The gel showed again these three bands (2.070 and 3.360 and ~4 Kb)
- => Colonies 8, 9 and 10 are not the desired construct.
Generation of Backbone pSB1C3-AraC-lacZ
Test Restriction Digest
In order find out why pSB6A1-AraC-lacZ shows wrong restriction pattern, parental pSB1C3-AraC-lacZ was restriction digested using KpnI as well as KpnI & BamHI.
| Reagent | Amount [µl] |
|---|---|
| pSB1C3-AraC-lacZ | 5 |
| Enzymes | 1.5 KpnI |
| Buffer NEB 3(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 33.5 |
| Expected band | 5,436 bp (linerized) |
| Reagent | Amount [µl] |
|---|---|
| pSB1C3-AraC-lacZ | 5 |
| Enzymes | 1.5 KpnI & 1.5 BamHI |
| Buffer NEB 3(10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 32 |
| Expected bands | 4.906 bp & 530 bp |
Result
Expected bands: 5.436 bp and 4.906 bp & 530 bp
- => pSB1C3-AraC(6)-lacZ is not desired construct. Pick new colony and test digest.
Miniprep
A new colony was picked from pSB1C3-AraC(6)-lacZ plate and a mini prep was performed.
Result
pSB1C3-AraC(6)-lacZ construct : 203 [ng/µl]
Test Restriction Digest
The new miniprep as well as one of the wrong pSB6A1-AraC(6)-lacZ ones were restriction digested with EcoRI-HF and PstI.
| Component | Amount [µl] |
|---|---|
| DNA | 3 |
| BSA (10x) | 5 |
| NEB2 buffer (10x) | 5 |
| Enzymes (EcoRI-HF and PstI) | 1.5 each |
| ddH2O | 34 |
- Incubated 1 h at 37°C
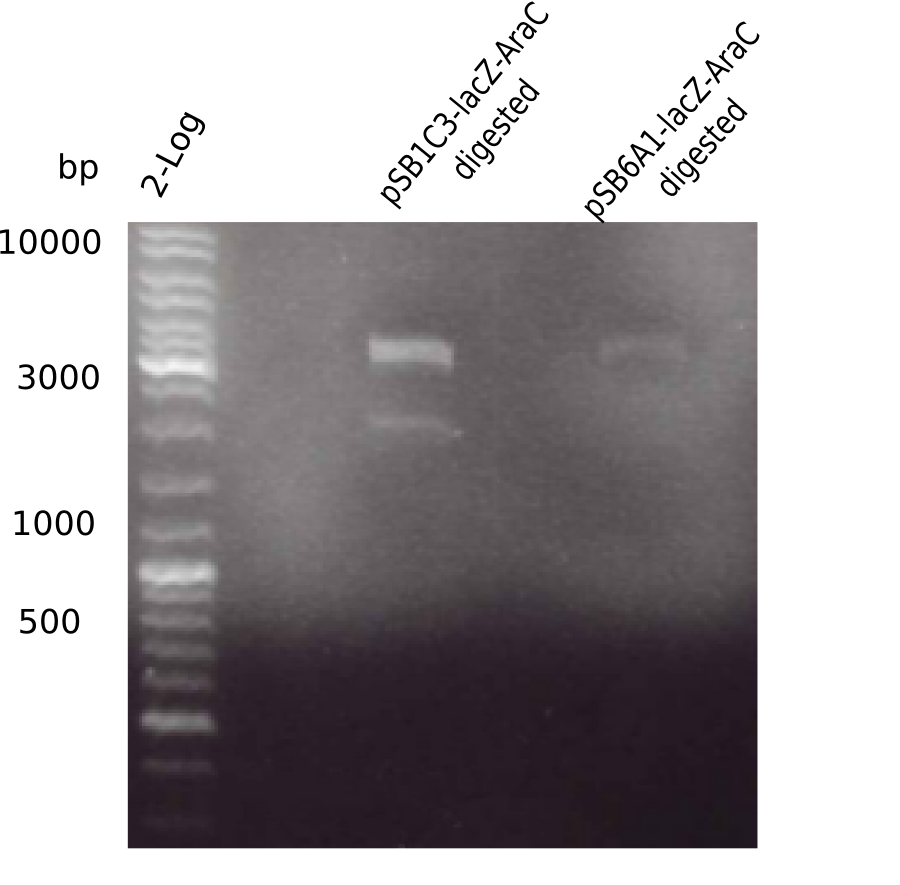
Result
Expected band: ? Kb
Gel showed again the three bands (2070 and 3360 and ~4 Kb).
- => pSB1C3-AraC(6)-lacZ is not desired construct. Start assembly all over again.
Test Restriction Digest pSB6A1-J04450
pSB6A1-J04450 was test digested to check for identity.
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-J04450 | 10 |
| Enzymes | 1.5 KpnI |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 28.5 |
| Expected band | 5,091 bp - Linearized |
| Reagent | Amount [µl] |
|---|---|
| pSB6A1-J04450 | 10 |
| Enzymes | 1.5 KpnI & 1.5 BamHI |
| Buffer NEB 3 (10x) | 5 |
| BSA (10x) | 5 |
| ddH2O | 27 |
| Expected bands | 4,906 bp & 530 bp |
Result
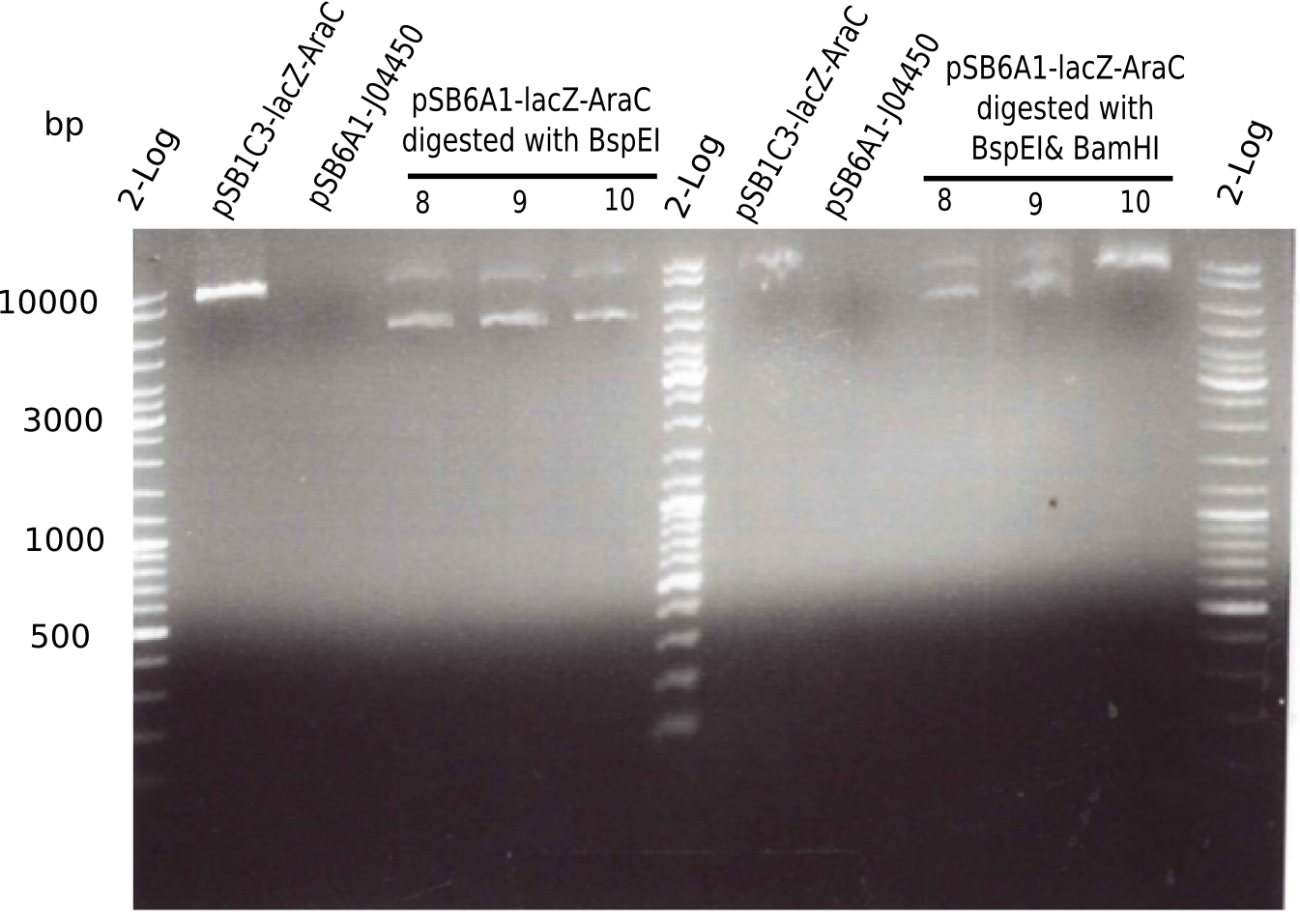
| Sample | Expected bands with KpnI | Bands on gel with KpnI | Expected bands with KpnI & BamHI | Bands on gel with KpnI & BamHI |
|---|---|---|---|---|
| pSB1C3-AraC-lacZ | 5.436 Kb (linerized) | 10 Kb | 4.906 Kb & 530 Kb | no band |
| pSB6A1-J04450 | 5.091 Kb (linearized) | no band | 5.091 Kb (linearized) | no band |
| pSB6A1-AraC-lacZ (8, 9, 10) | 7.388 Kb (linearized) | 6 Kb & 10 Kb | 4.607 Kb & 2.781 Kb | 6Kb & 10 Kb |
This means:
- The chloramphenicol backbone has the right length.
- The digested chloramphenicol backbone is also ok.
- The digested fragment pSB6A1-AraC-lacZ (with PstI & EcoRI-HF) showed the 2 expected bands at 2,070 bp and 3,360 bp, but another band around ~4 Kb.
- => Something is not ok with this ligated construct and we have to start all over again.
 "
"