Team:Heidelberg/Templates/Del week13 AE
From 2013.igem.org
(Difference between revisions)
(Created page with "==26-07-2013== ===Restriction digest of fragment FS_02 to FS_03; 5.3 kb; 08-07-2013 with EcoRI-HF=== [[File:20130726 2,5µllog2 AEEcoRIHF AFEcoRIHF GBGlII O...") |
|||
| Line 1: | Line 1: | ||
==26-07-2013== | ==26-07-2013== | ||
===Restriction digest of fragment FS_02 to FS_03; 5.3 kb; [[DelA-E#08-07-2013|08-07-2013]] with EcoRI-HF=== | ===Restriction digest of fragment FS_02 to FS_03; 5.3 kb; [[DelA-E#08-07-2013|08-07-2013]] with EcoRI-HF=== | ||
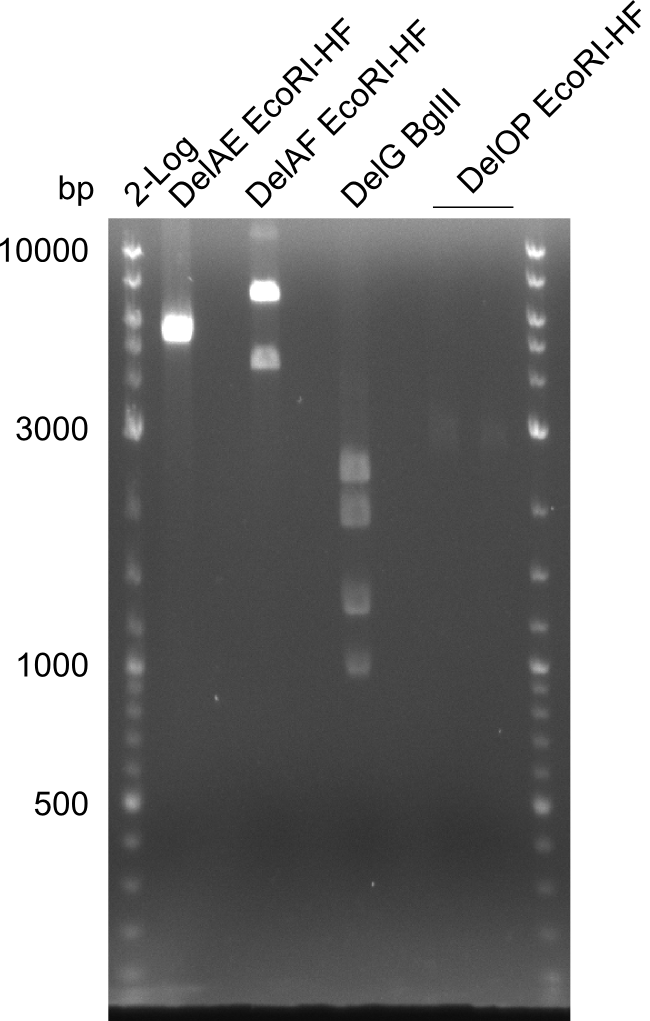
| - | [[File: | + | [[File:Heidelberg_20130726 2,5µllog2 AEEcoRIHF AFEcoRIHF GBGlII OPEcoRIHFbesch.png|100px|thumb|Restriction digest of DelAE (FS_02 to FS_03 ;08.07), DelAF (FS_02 to FS_05; 04.07) and DelOP (FS_22 to FS_13short ;21.07) with EcoRI-HF and digest of DelG (FS_08 to FS_23 cut1 ;23.07) with BglII; run at 100 V, 0.8 % gel (TAE)]] |
Incubation at 37°C for 1 h 45 min | Incubation at 37°C for 1 h 45 min | ||
| Line 24: | Line 24: | ||
==27-07-2013== | ==27-07-2013== | ||
===Amplificaction from FS_02 to FS_03; 5.3 kb=== | ===Amplificaction from FS_02 to FS_03; 5.3 kb=== | ||
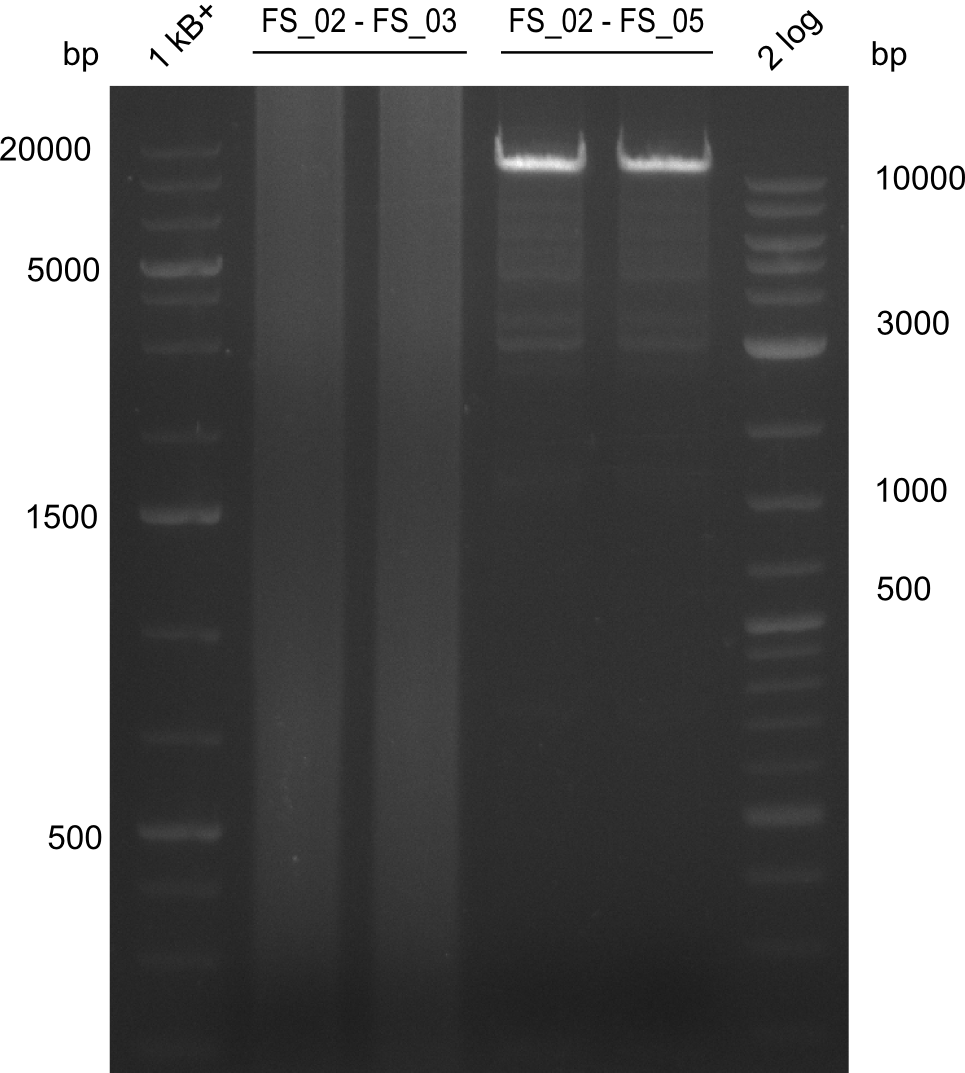
| - | [[File: | + | [[File:Heidelberg_20130727 2log 2xAE 2xAF 1kb 2xIliabesch.png|100px|thumb|Amplification of DelAE (FS02 to FS03; 27.07,1), Amplification of DelAF (FS_02 to FS05; 27.07); run at 100 V, 0.8 % gel (TAE)]] |
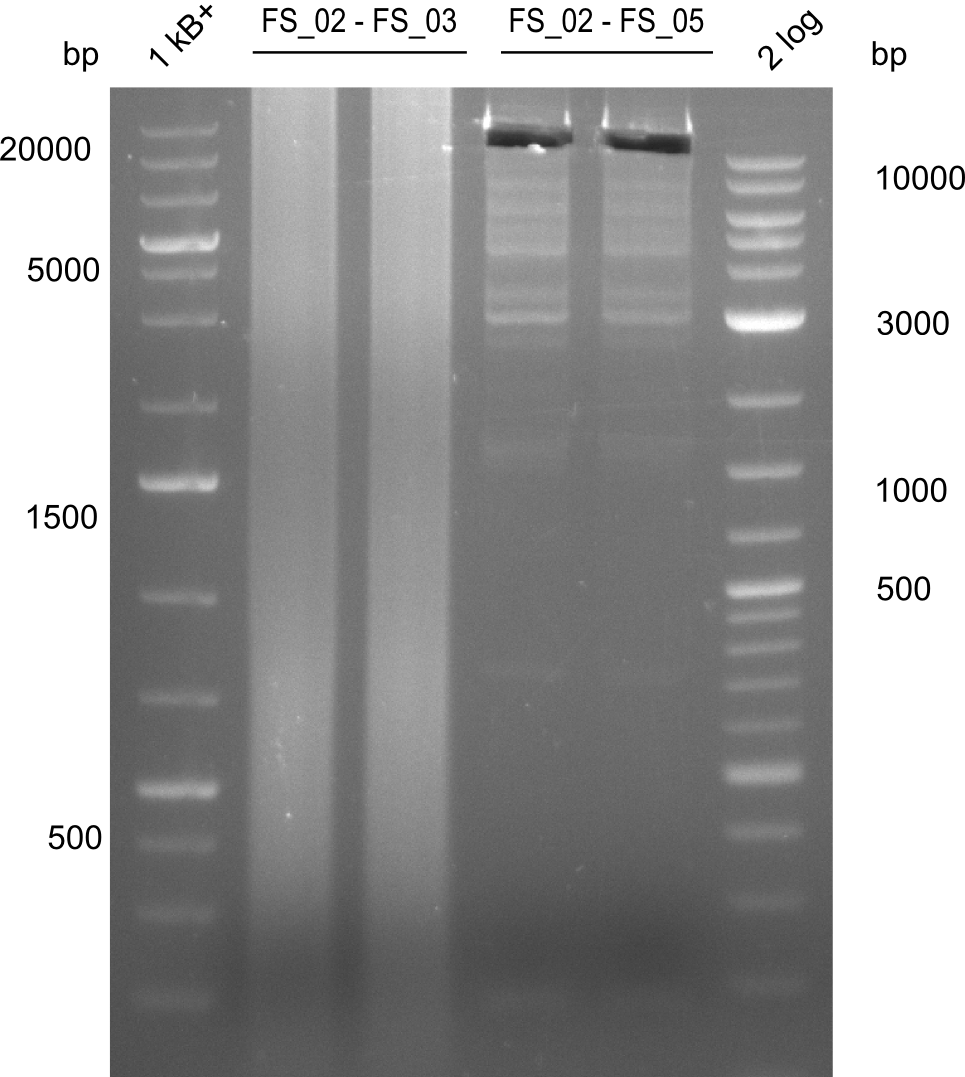
| - | [[File: | + | [[File:Heidelberg_20130727 2log 2xAE 2xAF 1kb 2xIliacut.png|100px|thumb|Amplification of DelAE (FS02 to FS03; 27.07,1), Amplification of DelAF (FS_02 to FS05; 27.07), cut; run at 100 V, 0.8 % gel (TAE)]] |
:'''Reaction''' | :'''Reaction''' | ||
| Line 32: | Line 32: | ||
! what !! µL | ! what !! µL | ||
|- | |- | ||
| - | | ''D. acidovorans'' || 1 | + | | ''D. acidovorans'' DSM-39 || 1 |
|- | |- | ||
| FS_02: (1/10) || 2.5 | | FS_02: (1/10) || 2.5 | ||
| Line 77: | Line 77: | ||
===Amplificaction from FS_02 to FS_03; 5.3 kb=== | ===Amplificaction from FS_02 to FS_03; 5.3 kb=== | ||
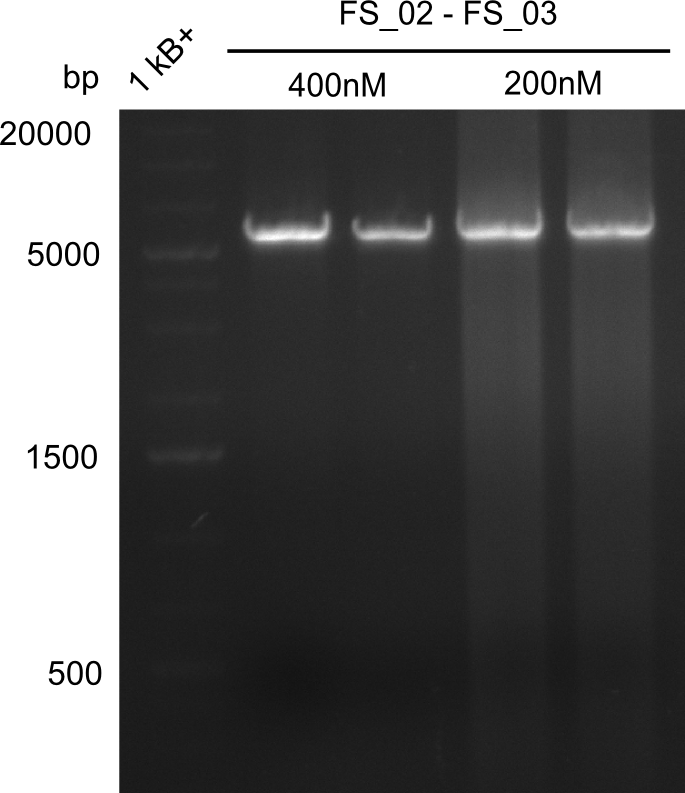
| - | [[File: | + | [[File:Heidelberg_20130728 1kbladder 2x DelAE FS2to3 5u prim 2x DelAE FS2to3 2,5u prim.png|100px|thumb|2nd Amplification of DelAE (FS02 to FS03; 27.07,1), first two lanes with 5 µL primer, second two lanes with 2.5 µL primer; run at 100 V, 0.8 % gel (TAE)]] |
| - | [[File: | + | [[File:Heidelberg_20130728 1kbladder 2x DelAE FS2to3 5u prim 2x DelAE FS2to3 2,5u prim cut.png|100px|thumb| gel after excision]] |
:'''Reaction''' | :'''Reaction''' | ||
| Line 85: | Line 85: | ||
! what !! µL | ! what !! µL | ||
|- | |- | ||
| - | | ''D. acidovorans'' || 1.5/1 | + | | ''D. acidovorans'' DSM-39 || 1.5/1 |
|- | |- | ||
| FS_02: (1/10) || 2.5/5 | | FS_02: (1/10) || 2.5/5 | ||
| Line 129: | Line 129: | ||
===Amplification I from FS_02 to FS_24; 7.1 kb=== | ===Amplification I from FS_02 to FS_24; 7.1 kb=== | ||
| - | [[File: | + | [[File:Heidelberg_20130627 log2 1kbladder FS02toFS24,200µM68td FS02toFS24,400µM68td FS02toFS24,200µM65 FS02toFS24,400µM65 FS06toFS24 FS20toFS24.png|200px|thumb| Amplifikation of DelFG (FS02-FS24), ; run at 100 V, 0.8 % gel (TAE)]] |
4 reactions, 2 with 200nM Primers and 2 with 400nM Primers (both concentrations for each condition) | 4 reactions, 2 with 200nM Primers and 2 with 400nM Primers (both concentrations for each condition) | ||
:'''Reaction''' | :'''Reaction''' | ||
| Line 136: | Line 136: | ||
! what !! µl | ! what !! µl | ||
|- | |- | ||
| - | | ''D. acidovorans'' || 1 | + | | ''D. acidovorans'' DSM-39 || 1 |
|- | |- | ||
| FS_02: (1/10) || 4/2 | | FS_02: (1/10) || 4/2 | ||
| Line 202: | Line 202: | ||
===Amplification II from FS_02 to FS_24; 7.1 kb=== | ===Amplification II from FS_02 to FS_24; 7.1 kb=== | ||
| - | [[File: | + | [[File:Heidelberg_20130727 2log 1kb 2-24.200nM.68td 2-24.400nM.68td 2-23.200nM.68td 2-23.400nM.68td 2-23.200nM.65 2-23.400nM.65.png|150px|thumb| 2 x Amplification of FS02 to FS_24 27.07.13 68°C Touchdown 200nM/400nM; 4x Amplification of FS-02 to FS23 27.0713 1 & 2 68°C Touchdown 200nM/400nM; 3. & 4. 65°C const. 200/400 nM, run at 100V ]] |
| Line 212: | Line 212: | ||
! what !! µl | ! what !! µl | ||
|- | |- | ||
| - | | ''D. acidovorans'' || 1 | + | | ''D. acidovorans'' DSM-39 || 1 |
|- | |- | ||
| FS_02: (1/10) || 2/4 | | FS_02: (1/10) || 2/4 | ||
| Line 278: | Line 278: | ||
===Amplification III from FS_02 to FS_24; 7.1 kb=== | ===Amplification III from FS_02 to FS_24; 7.1 kb=== | ||
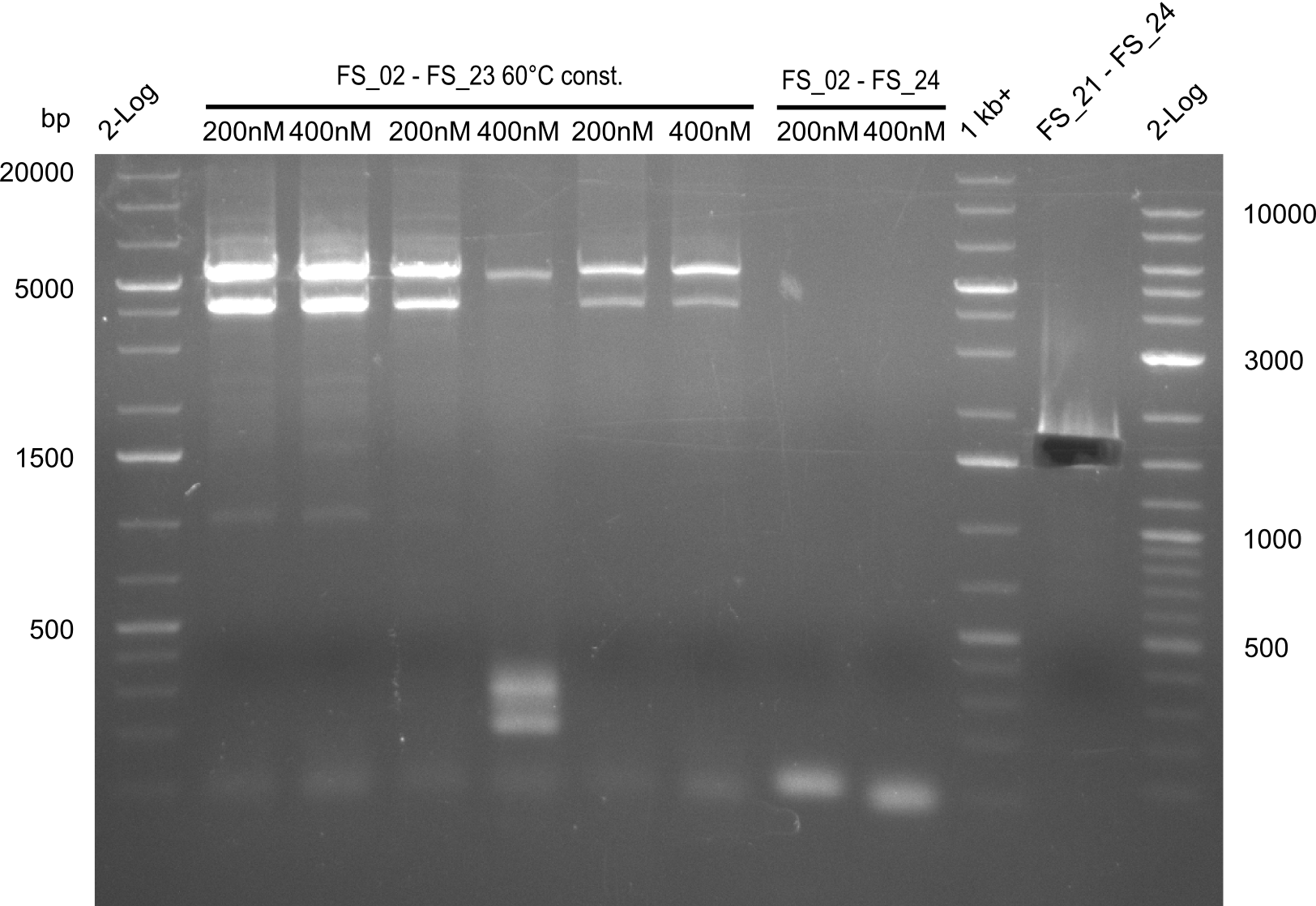
| - | [[File: | + | [[File:Heidelberg_20130728 1kbruler 4xFS02-FS23.60const.@T100.200nM T100.400nM old.200nM old.400nM red.200nM red.400nM FS02-FS24.janusA.200nM.66td FS02-FS24.T100.400nM.60const 1kb FS04-FS24.png|150px|thumb| 1kbruler / 4xFS02-FS23 60const @T100 200nM; T100 400nM; old 200nM; old 400nM ; red 200nM; red 400nM / FS02-FS24 66td janusA 200nM / FS02-FS24 T100 60const 200nM; 400nM 1kb FS04-FS24.png ; run at 100 V, 0.8 % gel (TAE)]] |
| - | [[File: | + | [[File:Heidelberg_20130728 1kbruler 4xFS02-FS23.60const.@T100.200nM T100.400nM old.200nM old.400nM red.200nM red.400nM FS02-FS24.janusA.200nM.66td FS02-FS24.T100.400nM.60const 1kb FS04-FS24 cut.png|150px|thumb| 1kbruler / 4xFS02-FS23 60const @T100 200nM; T100 400nM; old 200nM; old 400nM ; red 200nM; red 400nM / FS02-FS24 66td janusA 200nM / FS02-FS24 T100 60const 200nM; 400nM 1kb FS04-FS24.png ; run at 100 V, 0.8 % gel (TAE)]] |
2 reactions, 66°C Touchdown with 200nM Primers and 60°C constant with 400nM Primers | 2 reactions, 66°C Touchdown with 200nM Primers and 60°C constant with 400nM Primers | ||
| Line 288: | Line 288: | ||
! what !! µl | ! what !! µl | ||
|- | |- | ||
| - | | ''D. acidovorans'' || 1 | + | | ''D. acidovorans'' DSM-39 || 1 |
|- | |- | ||
| FS_02: (1/10) || 2/4 | | FS_02: (1/10) || 2/4 | ||
| Line 354: | Line 354: | ||
==29-07-2013== | ==29-07-2013== | ||
===Restriction digest of FS_02 to FS_03; 5.3 kb;([[DelA-E#27-07-2013|27-07-2013]]; II) with BglII === | ===Restriction digest of FS_02 to FS_03; 5.3 kb;([[DelA-E#27-07-2013|27-07-2013]]; II) with BglII === | ||
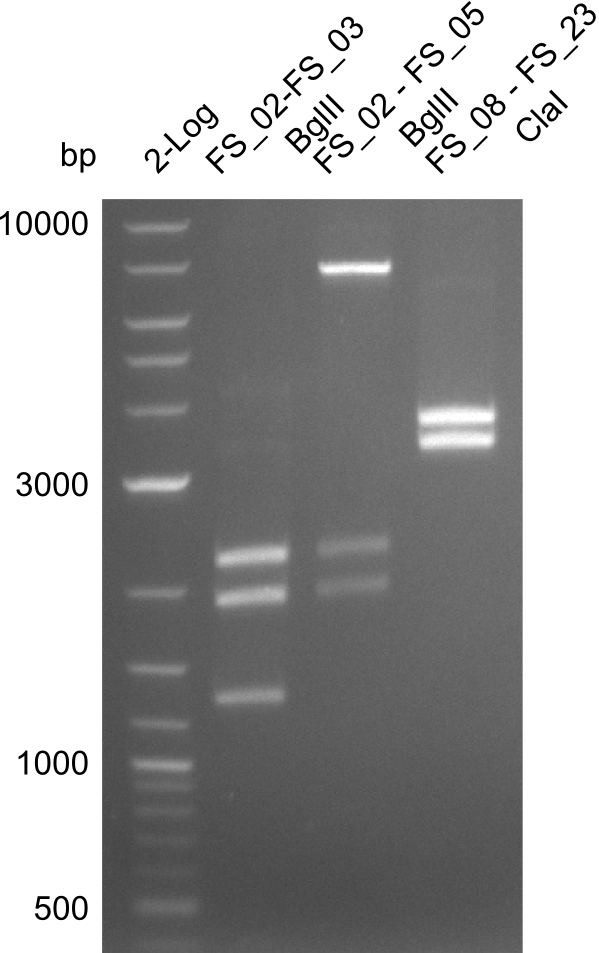
| - | [[File: | + | [[File:Heidelberg_20130729restrictiondigest 2log FS02-FS03 FS02-FS05 FS08-FS23besch.png|100px|thumb|Test restriction digest (29.07); run at 100 V, 0.8 % gel (TAE)]] |
Incubation at 37°C for about 3 h | Incubation at 37°C for about 3 h | ||
| Line 370: | Line 370: | ||
|} | |} | ||
Expected fragment lengths: 2,146 kb; 1,862 kb; 1,306 kb | Expected fragment lengths: 2,146 kb; 1,862 kb; 1,306 kb | ||
| - | + | ||
'''Results:''' | '''Results:''' | ||
* Restriction digest shows the expected product sizes | * Restriction digest shows the expected product sizes | ||
* indicator for correct amplicon but to be sure, PCR product will be prepared for single read sequencing by GATC | * indicator for correct amplicon but to be sure, PCR product will be prepared for single read sequencing by GATC | ||
Revision as of 01:36, 30 September 2013
Contents |
26-07-2013
Restriction digest of fragment FS_02 to FS_03; 5.3 kb; 08-07-2013 with EcoRI-HF
Incubation at 37°C for 1 h 45 min
| what | µL |
|---|---|
| FS_02 to FS_03 (08-07-2013) | 15 |
| EcorRI-HF | 0.5 |
| Buffer CutSmart | 2 |
| dd H2O | 2 |
Expected fragment lengths: 3054 bp, 2260 bp
Results:
- restriction digest of DelAE did not work, since incubation time might have been to short
27-07-2013
Amplificaction from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2.5 |
| FS_03: (1/10) | 2.5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 19 |
- Conditions
| Biorad MyCycler | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 12 min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE didnt work out, only smear occured
- repeat PCR with better cycler
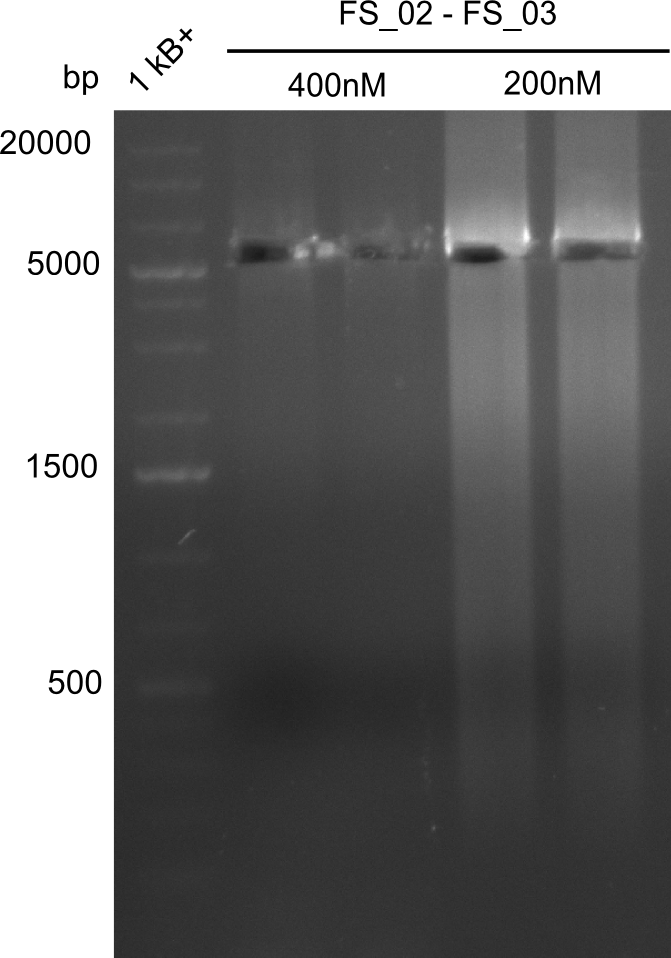
Amplificaction from FS_02 to FS_03; 5.3 kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans DSM-39 | 1.5/1 |
| FS_02: (1/10) | 2.5/5 |
| FS_03: (1/10) | 2.5/5 |
| Phusion flash Master Mix | 25 |
| DMSO | 2.5 |
| dd H2O | 19/14 |
- Conditions
| Biorad MyCycler* | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 2:30 | |
| 18 | 98 | 1 |
| 63 | 5 | |
| 72 | 2:30 | |
| 1 | 72 | 12 min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE worked with both 200 and 400 nM of Primers, nevertheless amplification was more specific with the higher primer concentration
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
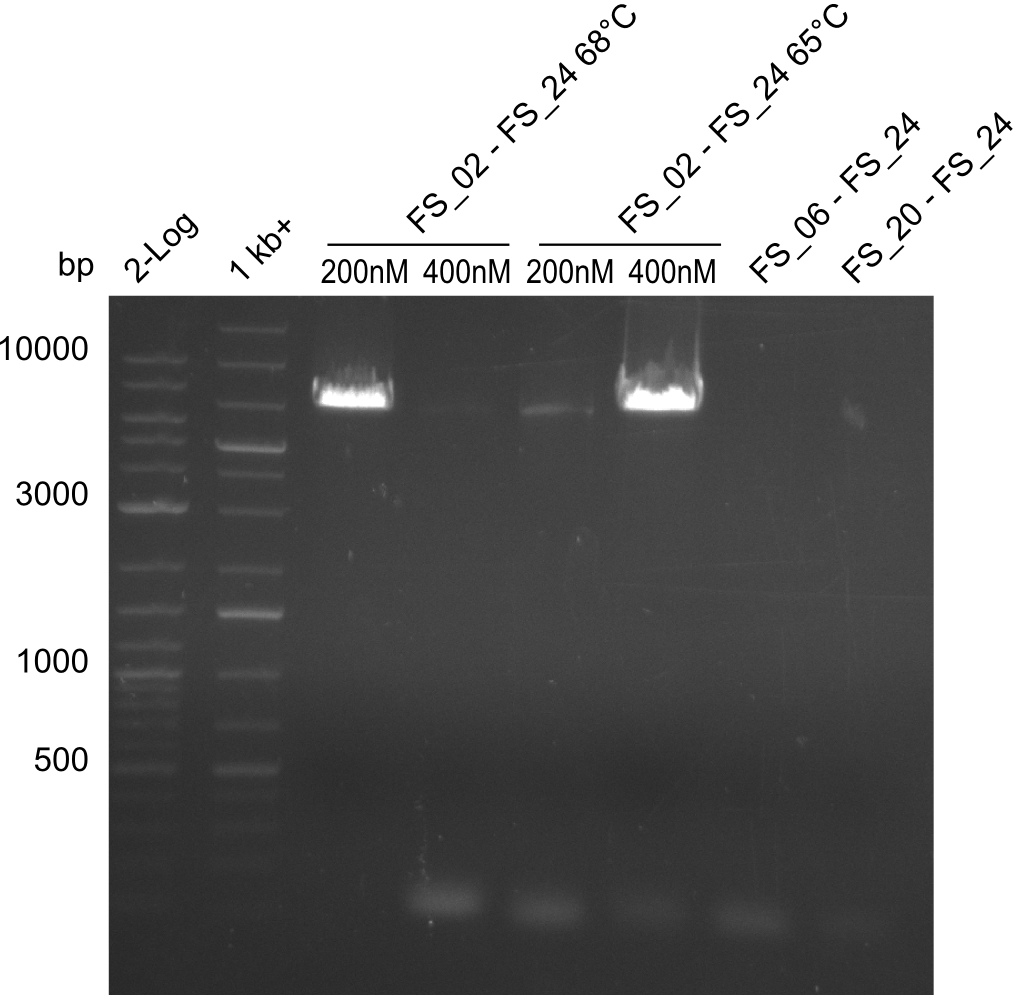
Amplification I from FS_02 to FS_24; 7.1 kb
4 reactions, 2 with 200nM Primers and 2 with 400nM Primers (both concentrations for each condition)
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 4/2 |
| FS_24: (1/10) | 4/2 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 3:50 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 3:50 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 3:50 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification of DelAE worked with 200 nM primer concentration at an annealing temperature of 68°C and 400 nM at an annealing temperature of 65°C, the product obtained at 65°C was more specific
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
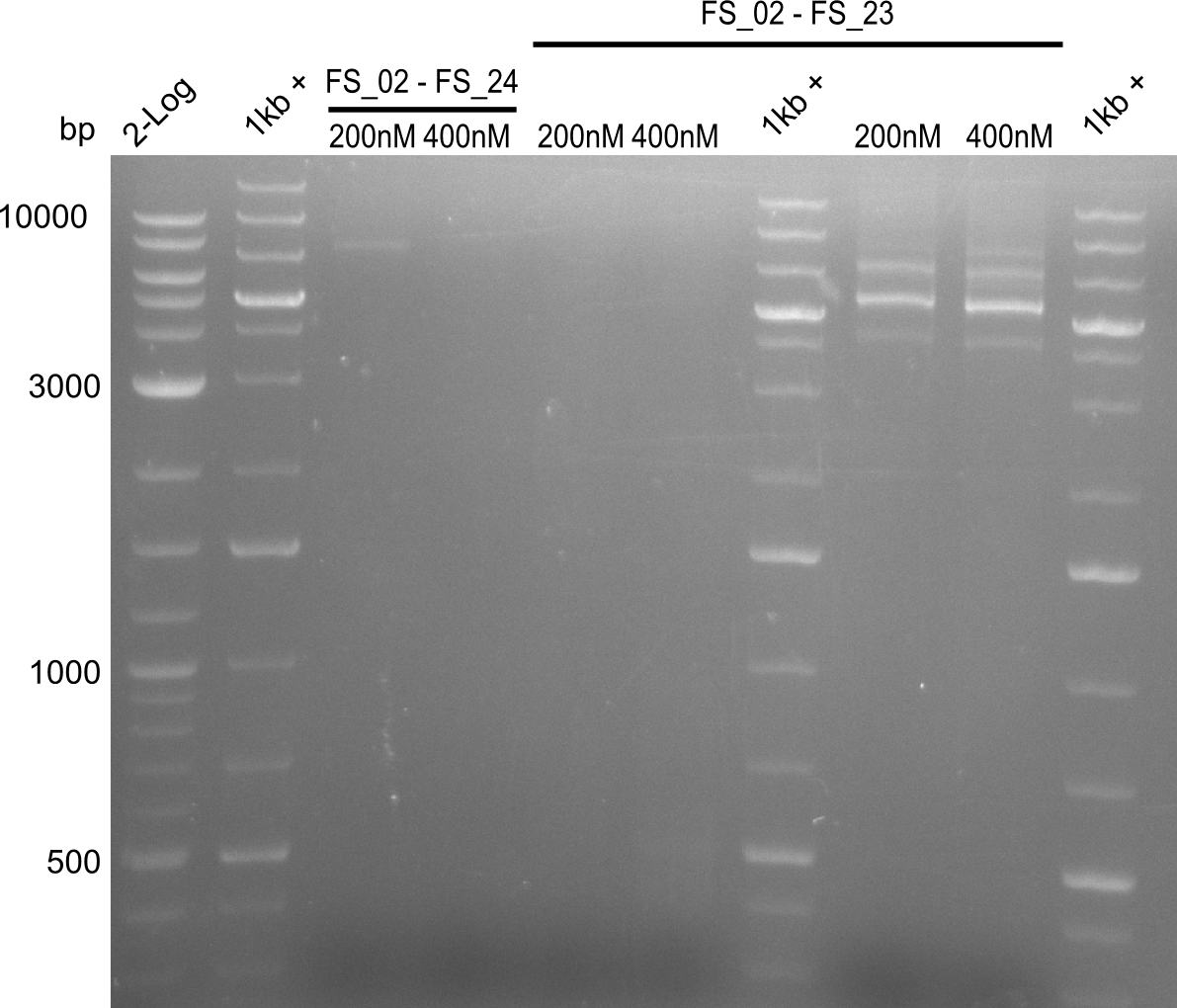
Amplification II from FS_02 to FS_24; 7.1 kb
2 reactions, 68°C Touchdown with 200nM Primers and 65°C constant with 400nM Primers
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2/4 |
| FS_24: (1/10) | 2/4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 65 | 5 | |
| 72 | 5:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 68 ↓ 0.5 | 5 | |
| 72 | 5:40 min | |
| 18 | 98 | 1 |
| 66 | 5 | |
| 72 | 5:40 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification did not work, neither with 200nM and 68°C touchdown, nor with 400nM and 65°C constant.
- Repeat amplfication with different conditions as primers did not bind very effectively
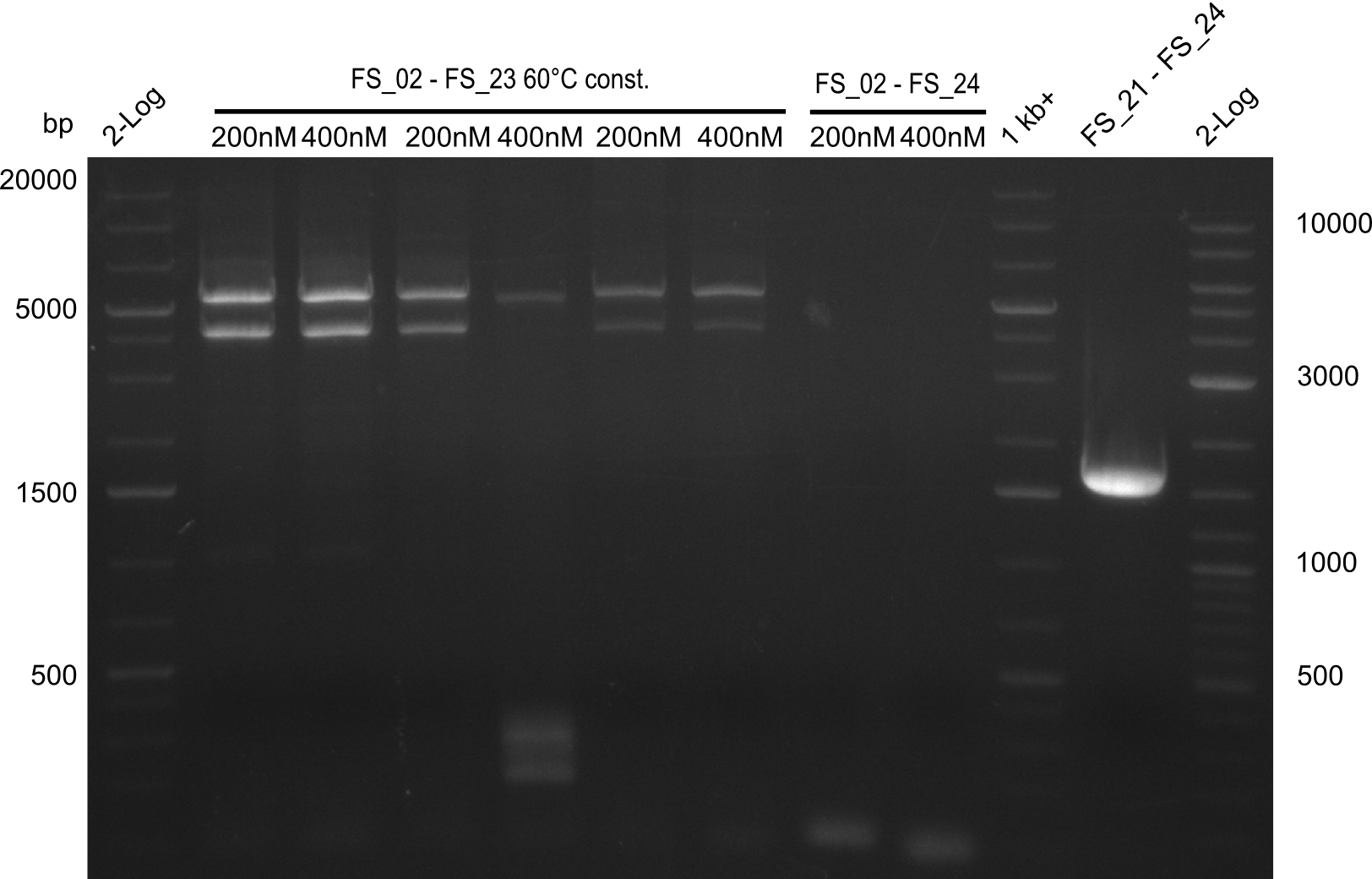
Amplification III from FS_02 to FS_24; 7.1 kb
2 reactions, 66°C Touchdown with 200nM Primers and 60°C constant with 400nM Primers
- Reaction
| what | µl |
|---|---|
| D. acidovorans DSM-39 | 1 |
| FS_02: (1/10) | 2/4 |
| FS_24: (1/10) | 2/4 |
| Phusion flash Master Mix | 10 |
| DMSO | 1 |
| dd H2O | 0/4 |
- Conditions I
| Biorad T100 | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 60 | 5 | |
| 72 | 5:40 | |
| 1 | 72 | 12 min |
| 1 | 10 | inf |
- Conditions II
| Biorad C1000 Touch Block A | ||
|---|---|---|
| Cycles-PCR | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| 66 ↓ 0.5 | 5 | |
| 72 | 5:40 min | |
| 18 | 98 | 1 |
| 64 | 5 | |
| 72 | 5:40 min | |
| 1 | 72 | 10 min |
| 1 | 4 | inf |
Results:
- Amplification from DelAE (7.1 kbp) failed again
- stick to the old strategy and use previously obtained fragments with different other fragments for gibson assembly.
29-07-2013
Restriction digest of FS_02 to FS_03; 5.3 kb;(27-07-2013; II) with BglII
Incubation at 37°C for about 3 h
| what | µL |
|---|---|
| FS_02 to FS_03 (27-07-2013; II) | 15 |
| BglII | 1 |
| Buffer 3.1 | 2 |
| dd H2O | 2 |
Expected fragment lengths: 2,146 kb; 1,862 kb; 1,306 kb
Results:
- Restriction digest shows the expected product sizes
- indicator for correct amplicon but to be sure, PCR product will be prepared for single read sequencing by GATC
 "
"