From 2013.igem.org
29-07-2013
Amplification from FS_21 to FS_24 ; (WRONG PRIMER!)
Amplifications of DelFG (29.07); run at 100 V, 0.8 % gel (TAE)
Amplifications of DelFG (29.07) cut
- Reaction
| what | µl
|
| D. acidovorans DSM-39 | 1
|
| FS_21: (1/10) | 2
|
| FS_24: (1/10) | 2
|
| Phusion flash Master Mix | 10
|
| DMSO | 1
|
| dd H2O | 4
|
- Conditions
| Biorad MyCycler*
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 30 | 98 | 1
|
| 55 | 5
|
| 72 | 2:30
|
| 1 | 72 | 8 min
|
| 1 | 12 | inf
|
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
Amplification from FS_21 to FS_24; (WRONG PRIMER!)
Amplification of DelFG (FS21 to FS26; 29.07); run at 100 V, 0.8 % gel (TAE)
Amplification of DelFG (FS21 to FS26; 29.07) cut
- Reaction
| what | µl
|
| D. acidovorans DSM-39 | 1
|
| FS_21: (1/10) | 2
|
| FS_24: (1/10) | 2
|
| Phusion flash Master Mix | 10
|
| DMSO | 1
|
| dd H2O | 4
|
- Conditions
| Biorad T100
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 30 | 98 | 1
|
| 58 | 5
|
| 72 | 2:30
|
| 1 | 72 | 8 min
|
| 1 | 12 | inf
|
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
Amplification from FS_21 to FS_24; (WRONG PRIMER!)

Amplification of DelFG (FS21 to FS26; 29.07); run at 100 V, 0.8 % gel (TAE)

Amplification of DelFG (FS21 to FS26; 29.07) cut
- Reaction
| what | µl
|
| D. acidovorans DSM-39 | 1
|
| FS_21: (1/10) | 2
|
| FS_24: (1/10) | 2
|
| Phusion flash Master Mix | 10
|
| DMSO | 1
|
| dd H2O | 4
|
- Conditions
| Biorad T100
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 35 | 98 | 1
|
| 60 | 5
|
| 72 | 2:30
|
| 1 | 72 | 8 min
|
| 1 | 12 | inf
|
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
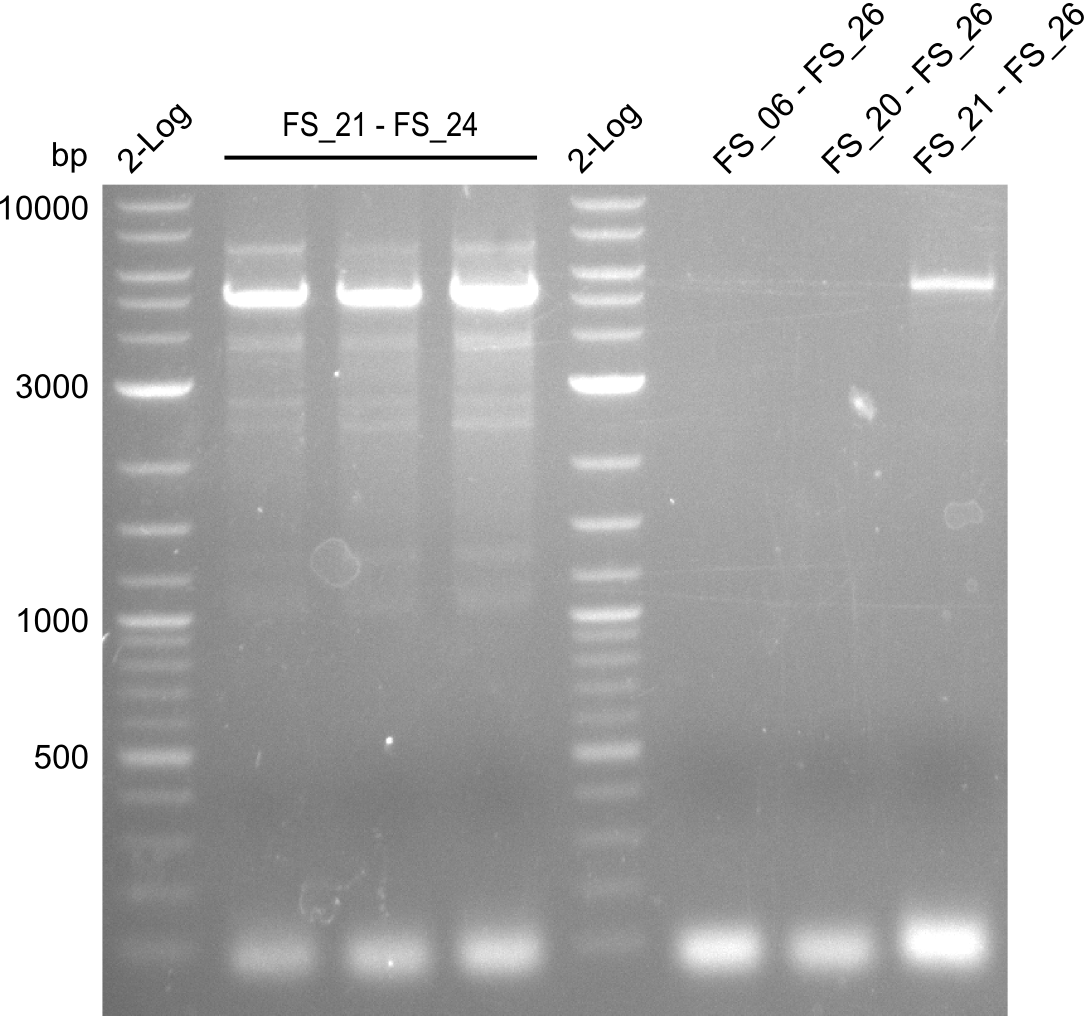
Amplification from FS_06/FS_20/FS_21 to FS_26 ; 5.5 kb

Amplifications of DelFG (29.07); run at 100 V, 0.8 % gel (TAE)

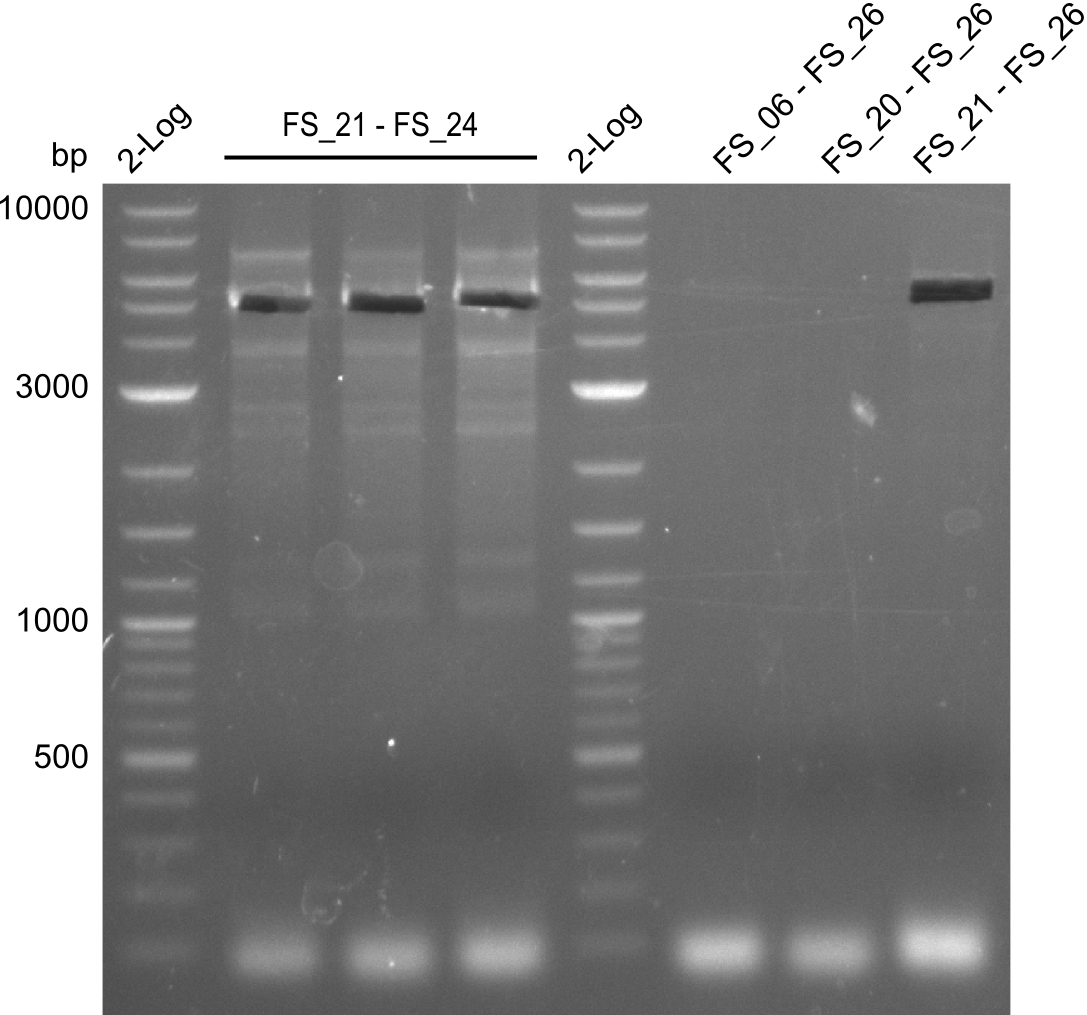
Amplifications of DelFG (29.07) cut
3x20µl
- Reaction
| what | µl
|
| D. acidovorans DSM-39 | 1
|
| FS_06 or FS_20 or FS_21: (1/10) | 2
|
| FS_26: (1/10) | 2
|
| Phusion flash Master Mix | 10
|
| DMSO | 1
|
| dd H2O | 4
|
- Conditions
| Biorad T100
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 30 | 98 | 1
|
| 64 | 5
|
| 72 | 2:15
|
| 1 | 72 | 10 min
|
| 1 | 10 | inf
|
Results:
- PCR product occured, though wrong primers were used, unspecific binding of primers in the genome of D. Acidovorans is the putative reason for this event
- Furthermore, amplification with FS_21 to FS_26 led to the intended product and satisfying specifity
- Amplification will be repeated at the same annealing temperature to obtain the amount of PCR product required for Gibson Assembly
- Amplfication will be repeated at lower annealing temperature to increase the yield
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
30-07-2013
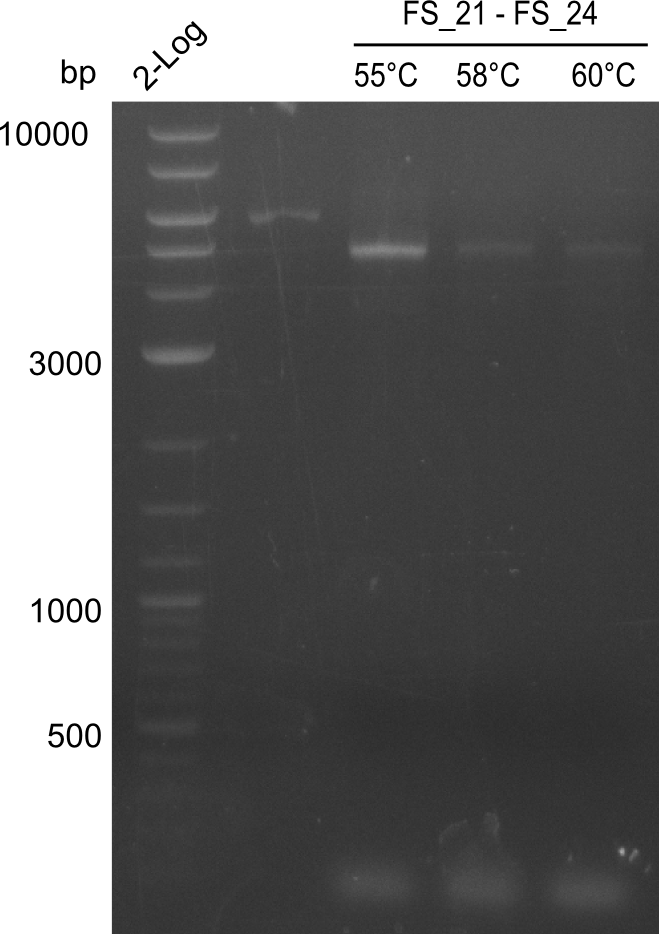
Amplification from FS_21 to FS_26 ; 5.5 kb
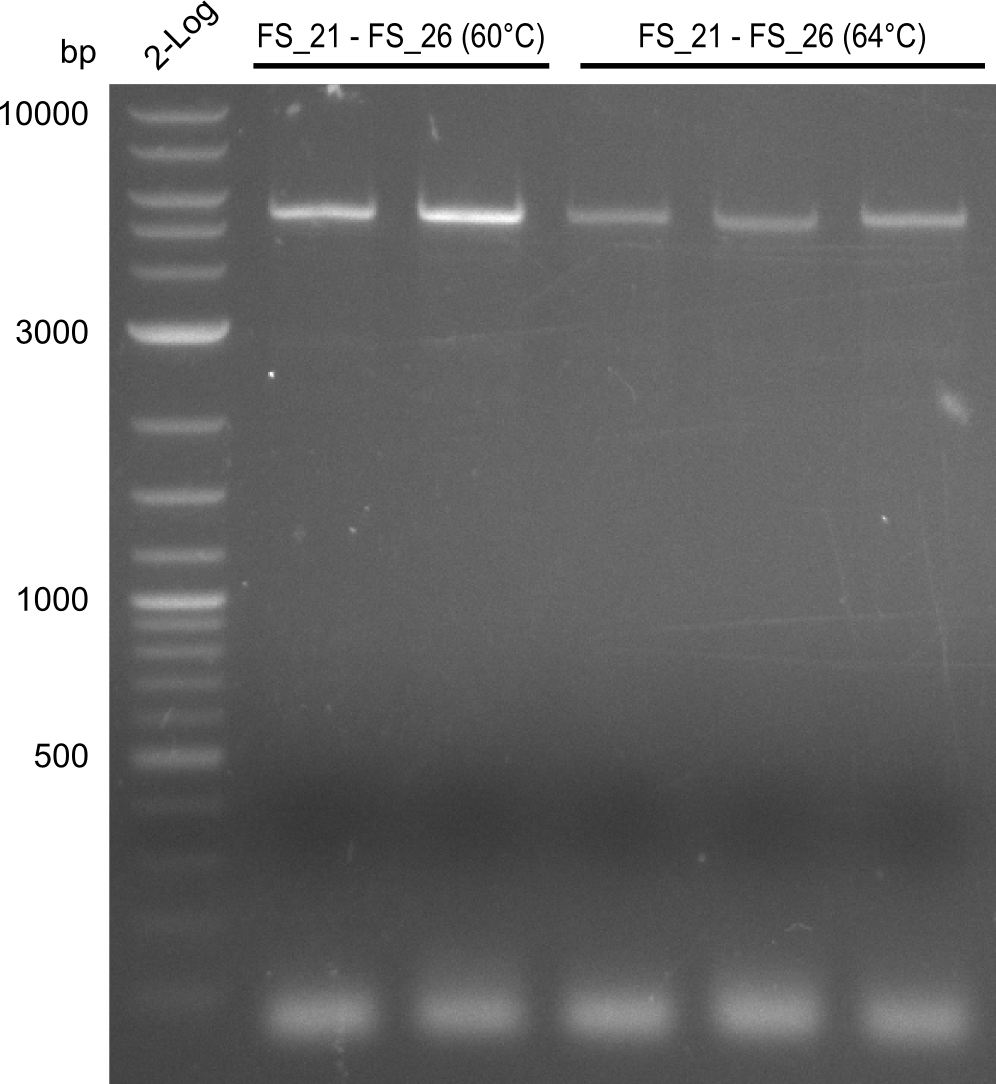
Amplification of DelFG (FS21 to FS26; 30.07) at 60°C and 64°C; run at 100 V, 0.8 % gel (TAE)
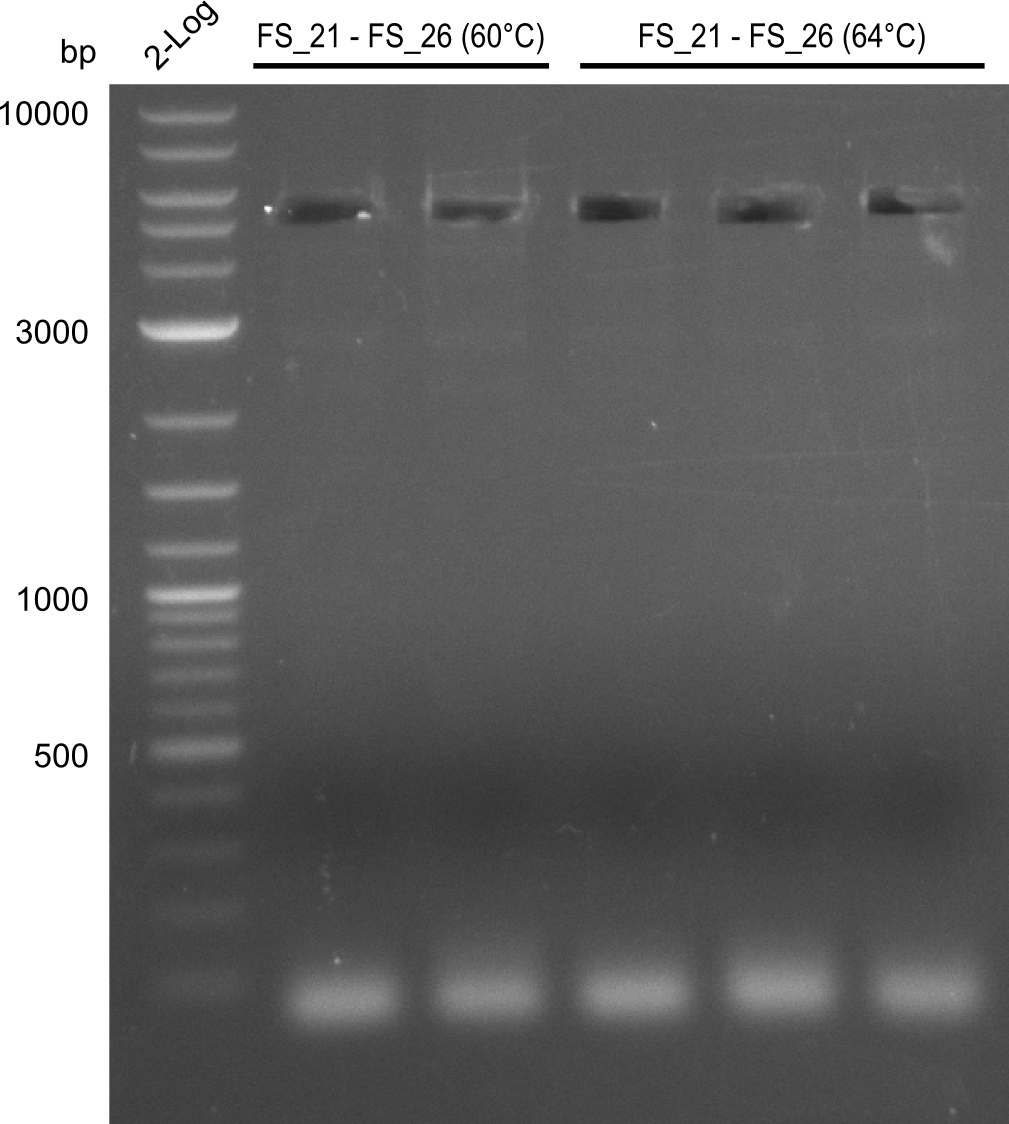
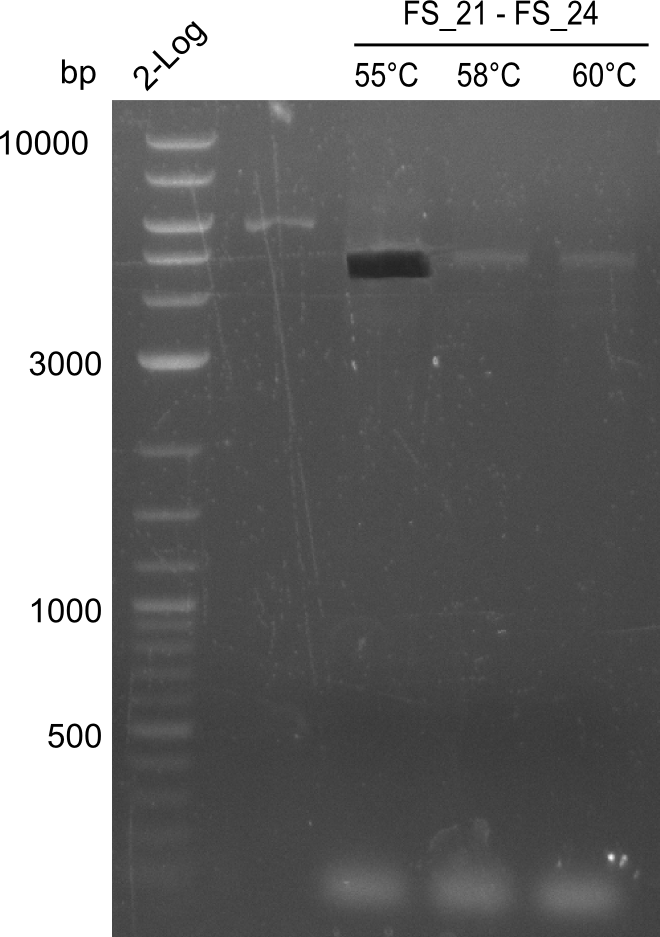
Amplification of DelFG (FS21 to FS26; 30.07) cut
3x20µl with conditions I, 2x20µl with conditions II
- Reaction
| what | µl
|
| D. acidovorans DSM-39 | 1
|
| FS_21: (1/10) | 2
|
| FS_26: (1/10) | 2
|
| Phusion flash Master Mix | 10
|
| DMSO | 1
|
| dd H2O | 4
|
- Conditions I
| Biorad T100
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 30 | 98 | 1
|
| 64 | 5
|
| 72 | 2:15
|
| 1 | 72 | 10 min
|
| 1 | 10 | inf
|
- Conditions II
| Biometra TProfessional Basic
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 30 | 98 | 1
|
| 60 | 5
|
| 72 | 2:15
|
| 1 | 72 | 10 min
|
| 1 | 10 | inf
|
Results:
- Amplification of DelFG was successful
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- restriction digest with XmaI will be conducted to validate the PCR product
31-07-2013
Restriction digest of fragment from FS_21 to FS_26; 5.5 kb; 29-07-2013) with XmaI
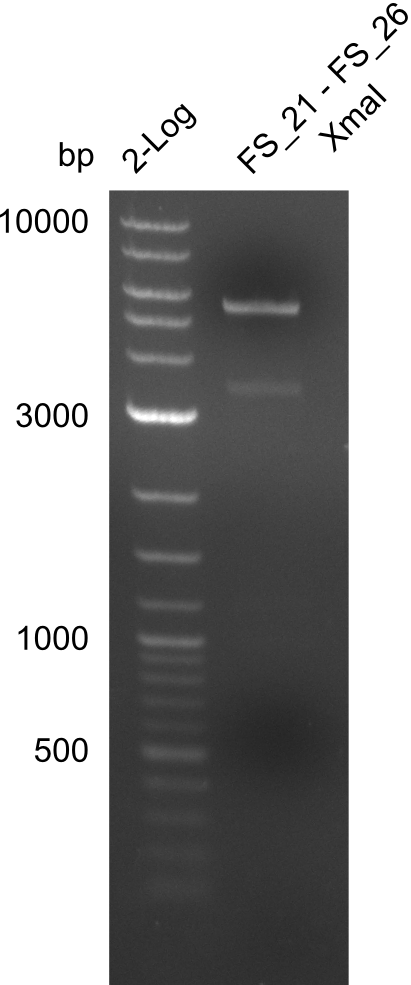
Restriction digest of FS21 to FS26 (29.7) with XmaI; run at 100 V, 0.8 % gel (TAE)
Incubation at 37°C for about 3 hours
| what | µl
|
| FS_21 to FS_26 (29-07-2013) | 17
|
| XmaI | 1
|
| Buffer CutSmart | 2
|
| Expected fragment lengths [bp] | 4268, 1202
|
Results:
- Restriction digest of DelFG with XmaI did not lead to the expected result
- digest will be carried out with a different enzyme, as the used one was outdated and digest might therefore not be very reliable
02-08-2013
Restriction digest of of fragment from FS_21 to FS_26; 5.5 kb; 30-07-2013) with ClaI
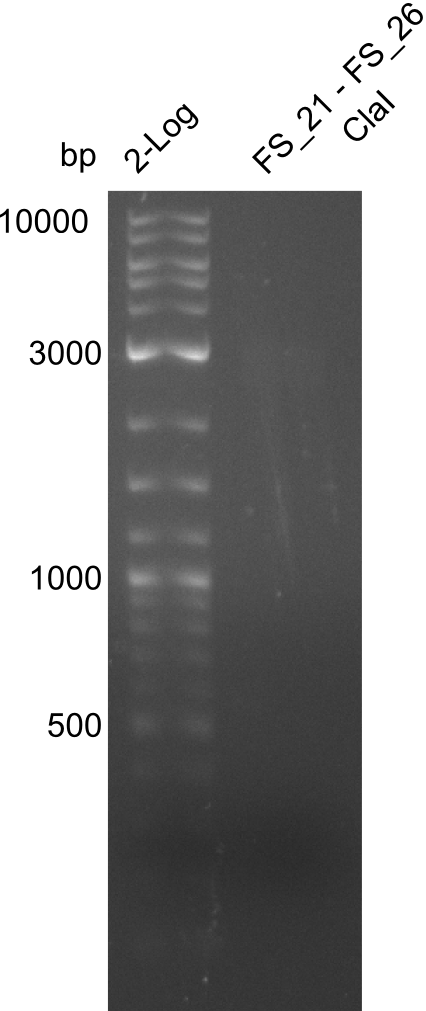
Restriction digest of FS21 to FS26 (30.7) with ClaI; run at 100 V, 0.8 % gel (TAE)
Incubation at 37°C for about 3 hours
| what | µl
|
| FS_21 to FS_26 (30-07-2013) | ~15
|
| ClaI | 1
|
| Buffer CutSmart | 2
|
| Expected fragment lengths [bp] | 2743, 1519, 1208
|
Results:
- Restriction digest of DelFG did not display any product
- digest will be repeated with higher amount of DNA after new amplification from the genome of D. Acidovorans
07-08-2013
Amplification from FS_21 to FS_26 ; 5.5 kb
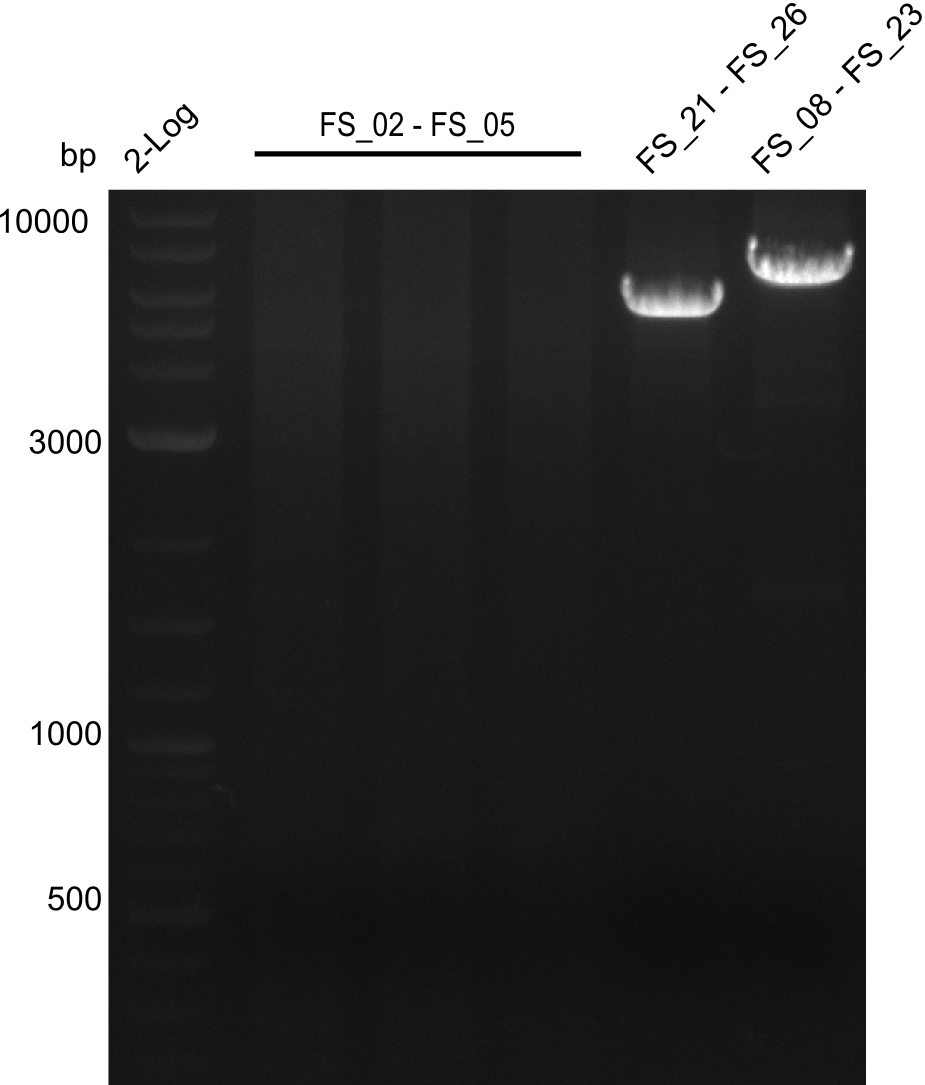
Amplifcation of DelAF(I), DelFG, DelG (07.08); run at 100 V, 0.8 % gel (TAE)

Amplifcation of DelAF(I), DelFG, DelG (07.08), cut
- Reaction
| what | µl
|
| D. acidovorans SPH-1 | 1
|
| FS_21: (1/10) | 2
|
| FS_26: (1/10) | 2
|
| Phusion flash Master Mix | 10
|
| DMSO | 1
|
| dd H2O | 4
|
- Conditions
| Biorad MyCycler
|
| Cycles | temperature [°C] | Time [s]
|
| 1 | 98 | 10
|
| 30 | 98 | 1
|
| 60 | 5
|
| 72 | 2:15
|
| 1 | 72 | 10 min
|
| 1 | 10 | inf
|
Results:
- Amplification of DelFG was successful, gel displays highly specific product of convincing yield
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- restriction digest with ClaI will be conducted to validate PCR product
 "
"









