Team:Heidelberg/Templates/Del week15 AE
From 2013.igem.org
(Difference between revisions)
| Line 7: | Line 7: | ||
<gallery> | <gallery> | ||
| - | File: | + | File:AlignementAE 02Sequenzierung.clustal.txt | Alignment of DelAE(FS_02 to FS_03) with Primer FS_02 |
| - | File: | + | File:AlignementAE 03Sequenzierung.clustal.txt | Alignment of DelAE(FS_02 to FS_03) with Primer FS_03 |
| - | + | ||
| - | + | ||
</gallery> | </gallery> | ||
Latest revision as of 19:17, 4 October 2013
Contents |
07-08-2013
Sequencing Results
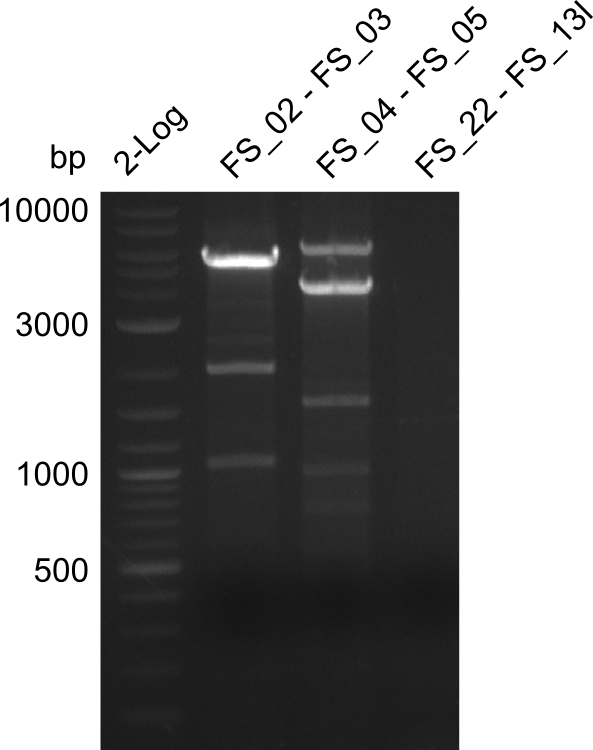
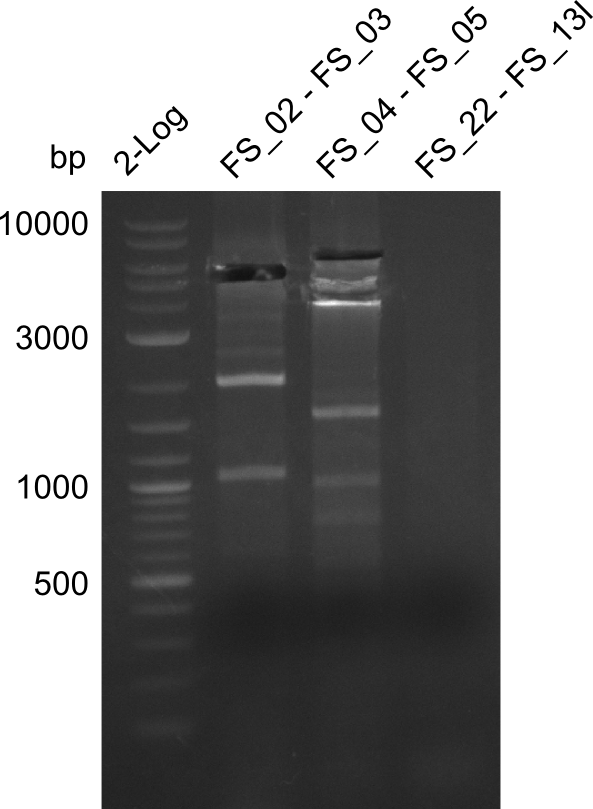
We send the following samples (amplified fragments of D. acidovorans DSM-39 and backbone pSB4K5 from the partsregistry) to GATC for sequencing:
- AE (FS_02-FS_03) with Primer FS_02
- AE (FS_02-FS_03) with Primer FS_03
08-08-2013
Amplification from FS_02 to FS_03; 5.3kb
- Reaction
| what | µL |
|---|---|
| D. acidovorans SPH-1 | 1 |
| FS_02: (1/10) | 4 |
| FS_03: (1/10) | 4 |
| Phusion flash Master Mix | 10 |
- Conditions
| Biometra TProfessional Basic | ||
|---|---|---|
| Cycles | temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 58 | 5 | |
| 72 | 1:30 | |
| 1 | 72 | 7min |
| 1 | 12 | inf |
Results:
- Amplification of DelAE was repeated successfully with the new strain SPH-1
- bands were cut out and DNA purified using QIAquick Gel Extraction Kit
- if fragment is used in Gibson assembly the amplification has to be repeated to increase the amount of product
 "
"