Team:Heidelberg/Templates/Delftibactin Results
From 2013.igem.org
| Line 18: | Line 18: | ||
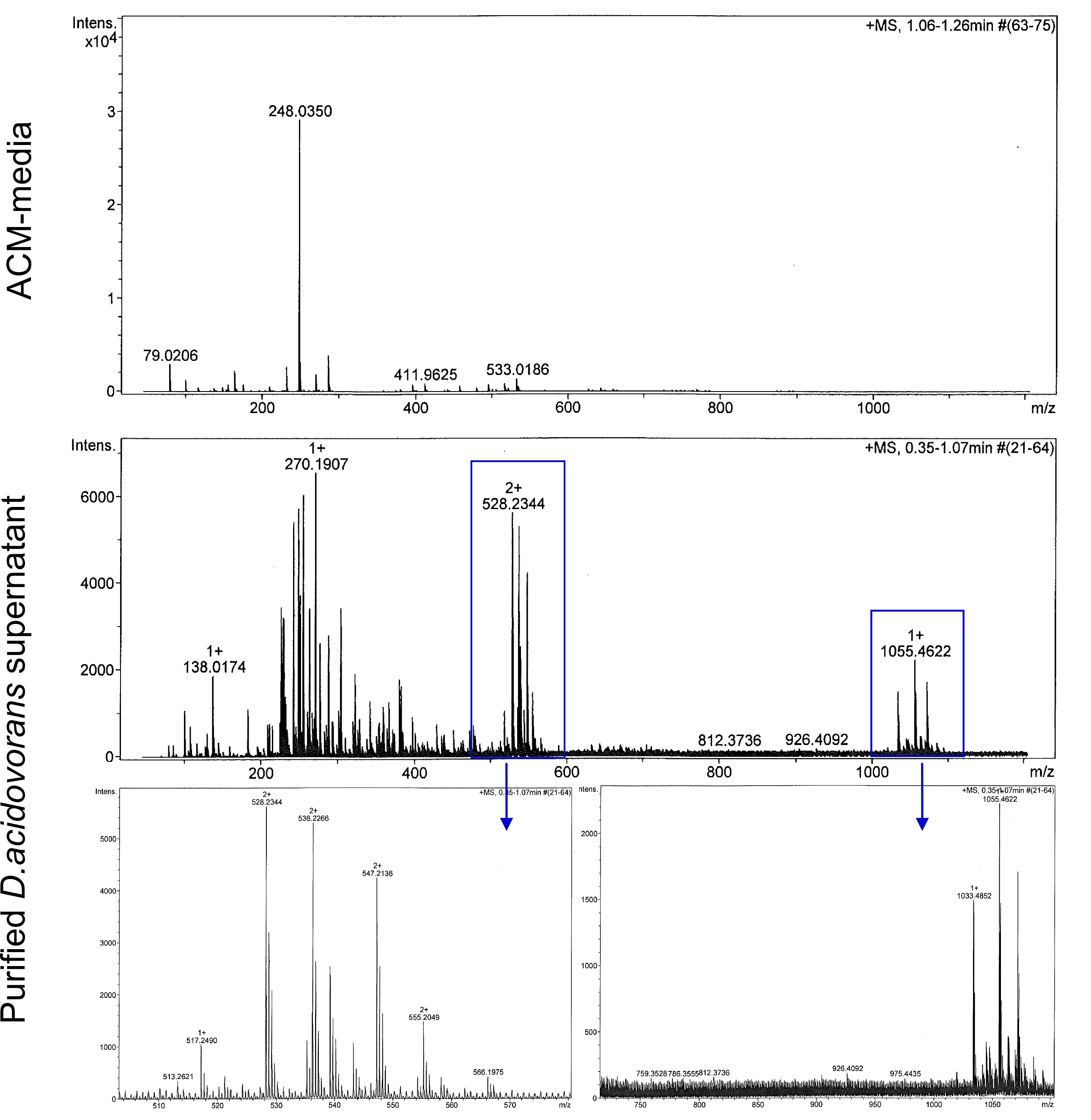
We confirmed purification of Delftibactin using HP20 resin. Additionally, we proved precipitation of gold by the purified Delftibactin (figures 6 and 7) and detected it by Micro-TOF. | We confirmed purification of Delftibactin using HP20 resin. Additionally, we proved precipitation of gold by the purified Delftibactin (figures 6 and 7) and detected it by Micro-TOF. | ||
| - | [[File:Heidelberg_Microtof-Delftia.png| | + | [[File:Heidelberg_Microtof-Delftia.png|400px]] |
<gallery widths="200px" heights="150px"> | <gallery widths="200px" heights="150px"> | ||
Revision as of 15:52, 4 October 2013
Producing Delftibactin
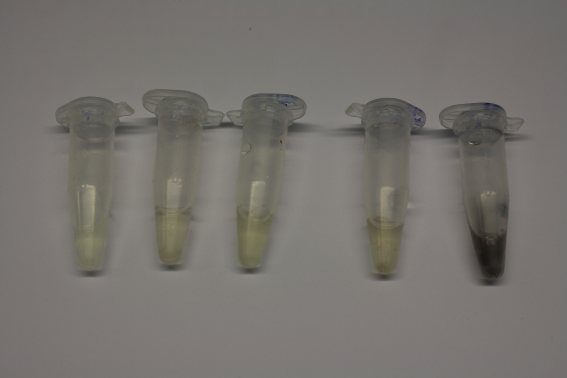
We obtained D. acidovorans DSM-39 from the ZSMZ and successfully reproduced the paper of Johnsson et al. (<bib id="pmid23377039"/>). In our experiments, precipitation on agar plates worked even better than described in the paper as shown in Figure 1. D. acidovorans is capable to precipitate solid gold from gold chloride solution as purple-black nanoparticles. Already at low concentrations of gold chloride, gold nonaparticles are precipitated increasing with concentration of gold chloride in solution (Fig. 2).
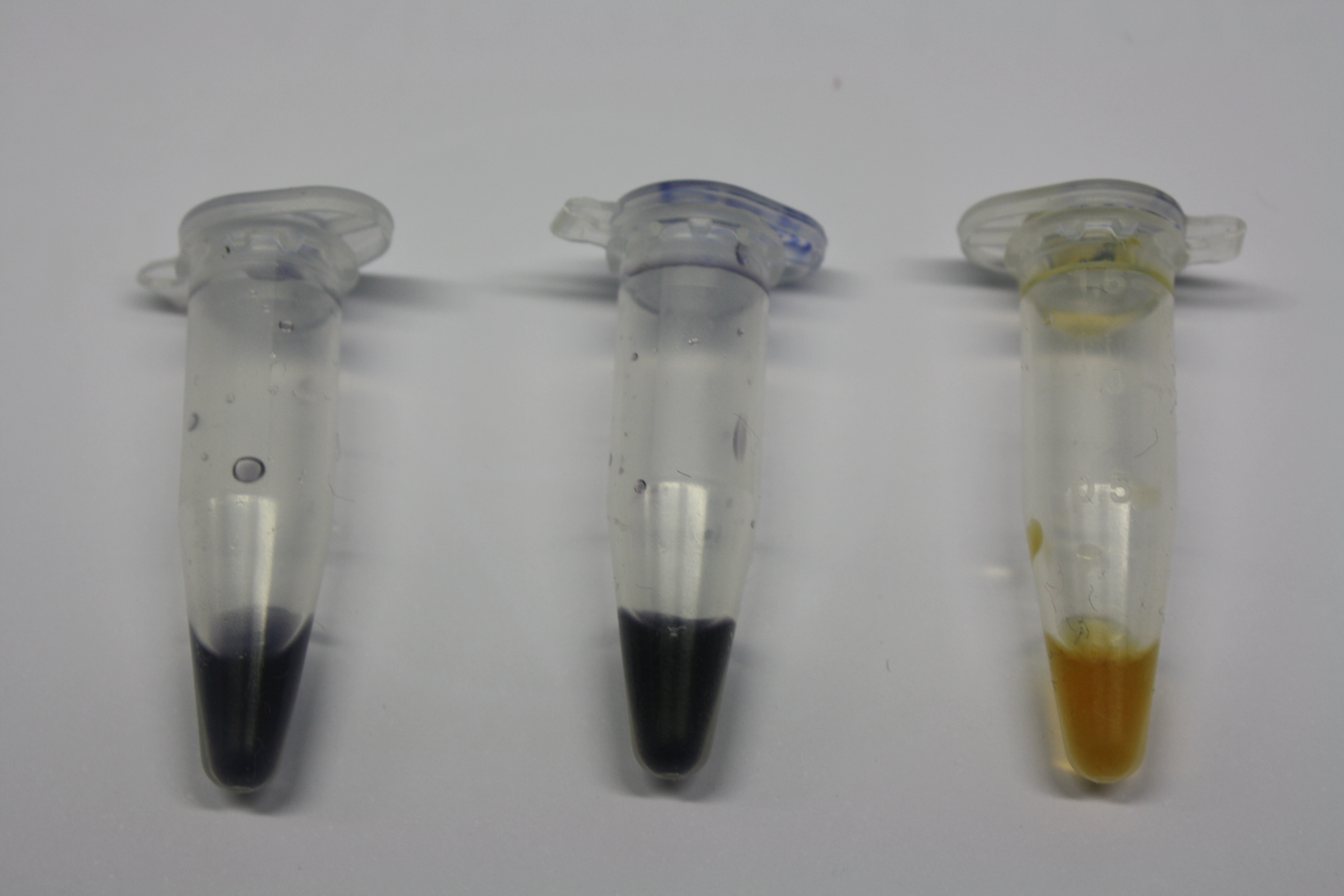
Using supernatants from the new Delftia acidovorans strain SPH-1, we showed precipitation of gold chloride solution to gold nanoparticles. Furthermore, we melted the purple-black nanoparticles to shiny solid gold as shown in figures 3 to 5.
We confirmed purification of Delftibactin using HP20 resin. Additionally, we proved precipitation of gold by the purified Delftibactin (figures 6 and 7) and detected it by Micro-TOF.
On the way to biological recycling by Delftibactin, we were able to dissolve gold-containing parts of an old CPU and established a protocol for recovery of gold as soluble gold salts from electronic waste (figures 8 to 10).
Amplification of the Del Cluster Genes
The first step towards introducing the Delftibactin pathway into E. coli was the amplification of the Del-cluster encoded on the genome of Delftia acidovorans. To this end, we designed Gibson primers and amplified the genes of the non-ribosomal peptide synthetase and the polyketide synthase (PKS) pathway as well as additional constitutive proteins, which were predicted to be necessary for the production of Delftibactin [1] . In the first weeks, PCRs were successfully established and optimized. At the same time, a separate plasmid was created encoding the PPTase from Bacillus subtilis and a permeability device [ BBa_I746200] [2]for the export of the synthesized NRP. Additionallly, plasmids from the partsregistry [3] were successfully transformed and amplified for the backbone generation.
Gibson Assembly & Transformation
As it is not trivial to assemble such a large number of long DNA fragments, we have used the Gibson Assembly method [4] implemented by Cambridge in iGEM 2010 instead of common cloning procedures such as restriction and ligation techniques. The assembled constructs of up to 32 kbp in size were transformed into E. coli via electroporation. Correct assembly of the fragments was tested by analytical restriction digest.
Figure 11: Restriction digest of three3 different plasmids needed for the NRPS/PKS pathway which to generated Delftibactin. a) Four4 digested colonies clones of the pIK8-plasmid, where clones 6 and 9 show the expected pattern. b) shows oneOne clone of E. coli clone with the 32 kpb DelRest -plasmid and was digested with three different enzymes and every lane shows the specific pattern for the according enzyme. c) Rshows the restriction digfest of the DelH -plasmid with PvuI. Clone 5 shows the expected pattern and is probably positive.
The exemplary restriction digest shown above (Fig. 11) confirms the correct assembly of the three desired constructs as it shows the expected band pattern expected from in silico digestion. Clones (6 and 9) contained the plasmid that encodes for the Methylmalonyl-CoA pathway (Fig. 11a) .The obtained DNA sequences were sent in for sequencing over the gibson-assembled regions for confirmation. [5][6] The sequencing confirmed the accuracy of the sequence. The successful generation of DelRest plasmid was proven by different enzymatic restriction digests (Fig. 11b) and also attested by the sequencing. The sequence was compared with the available SPH1 sequence of the Del cluster of Delftia acidovorans in NEB-database [7] (For further information please visit our labjournal).
After analytical restriction digest of DelH (Fig.11c), dozens of clones were positive. In co
 "
"