Team:Heidelberg/Templates/Indigoidine week17
From 2013.igem.org
Contents |
Preparation for T-Domain exchange
Fragment Amplification
| template: 0.5 ul from pRB19 miniprep, Primer KH3/4 | ||
| template: 0.5 ul from pRB19 miniprep, Primer KHRB21/KH4 | ||
| cycles | temperature (°C) | time (s) |
|---|---|---|
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 60 | 5 | |
| 72 | 100 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
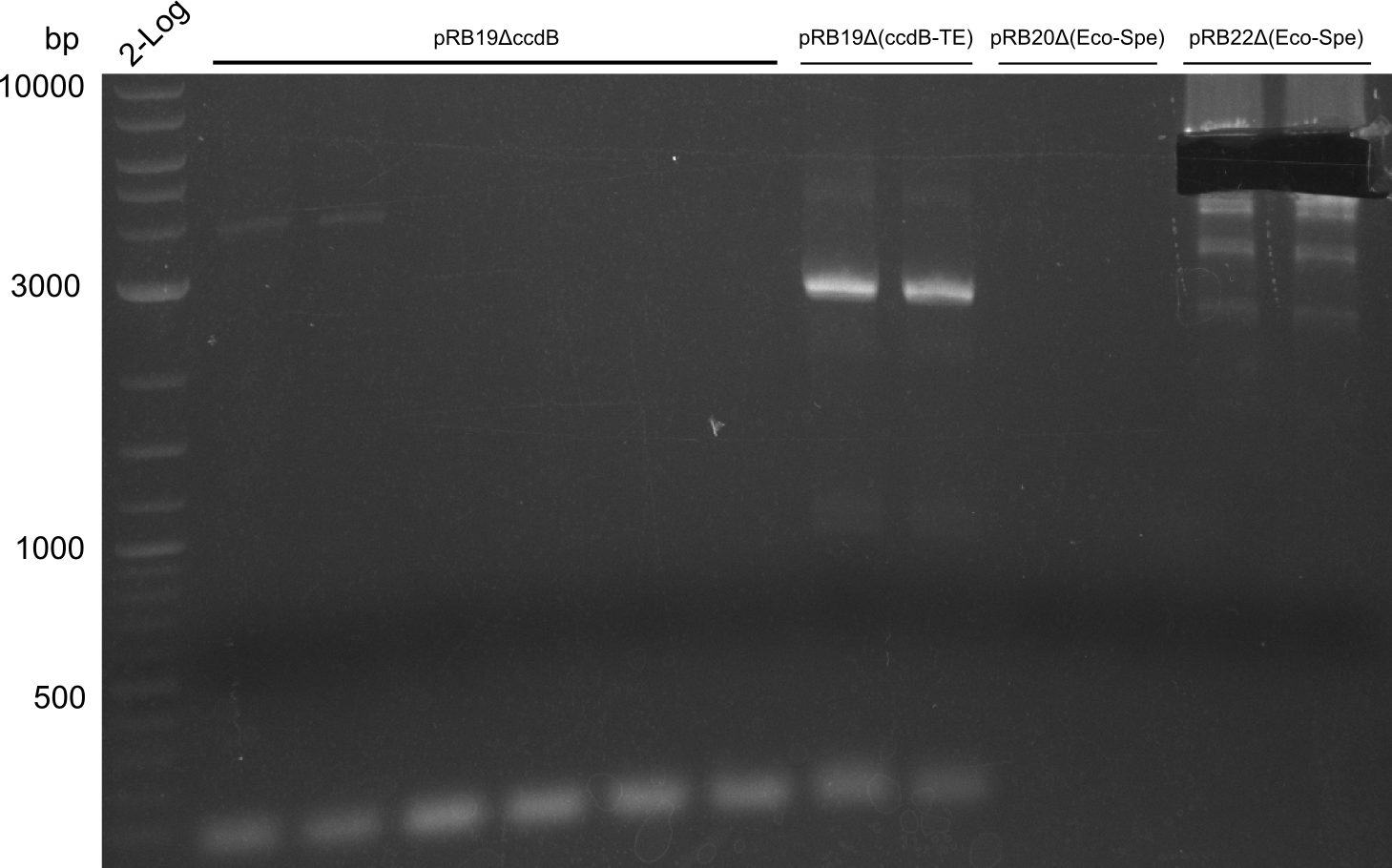
No band at 6400 bp. Second try with less stringent conditions
| template: 0.5 ul from pRB19 miniprep, Primer KH3/4 | ||
| template: 0.5 ul from pRB19 miniprep, Primer KHRB21/KH4 | ||
| cycles | temperature (°C) | time (s) |
|---|---|---|
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 62 ? 0.5 | 5 | |
| 72 | 100 | |
| 25 | 98 | 1 |
| 57 | 5 | |
| 72 | 100 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
Still nothing; maybe the MiniPrep didn't work so fine. We will screen pRB19-transformed cells on pRB19. We want to get a first impression on whether the Domain-exchange works, so we'll use pRB14 instead.
| template: 0.8 ug from pRB14 miniprep, Primer KH3/4 | ||
| template: 0.8 ug from pRB14 miniprep, Primer KHRB21/KH4 | ||
| cycles | temperature (°C) | time (s) |
|---|---|---|
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| 62 ? 0.5 | 5 | |
| 72 | 120 | |
| 25 | 98 | 1 |
| 57 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
PCR worked but with very low yield. We try to amplify from pRB19-transformed cells instead of the miniprep.
| template: 1 ul from pRB19-transformed cells, Primer KH3/4 | ||
| template: 1 ul from pRB19-transformed cells, Primer KHRB21/KH4 | ||
| cycles | temperature (°C) | time (s) |
|---|---|---|
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 57-60-68 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
25 ul Phusion HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul
| TTE-Domains in BioRAD cycler 2 | ||
|---|---|---|
| bpsA-TTE KH7/RB72 1 ng pMM64 miniprep | ||
| entF-TTE RB53/73 colony MG1655 | ||
| tycC6-TTE RB57/74 1 ul B. para overnight culture | ||
| delH5-TTE RB61/75 colony D. aci SPH-1 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 120 |
| 40 | 98 | 10 |
| 65 | 20 | |
| 72 | 30 | |
| 1 | 72 | 120 |
| 1 | 12 | - |
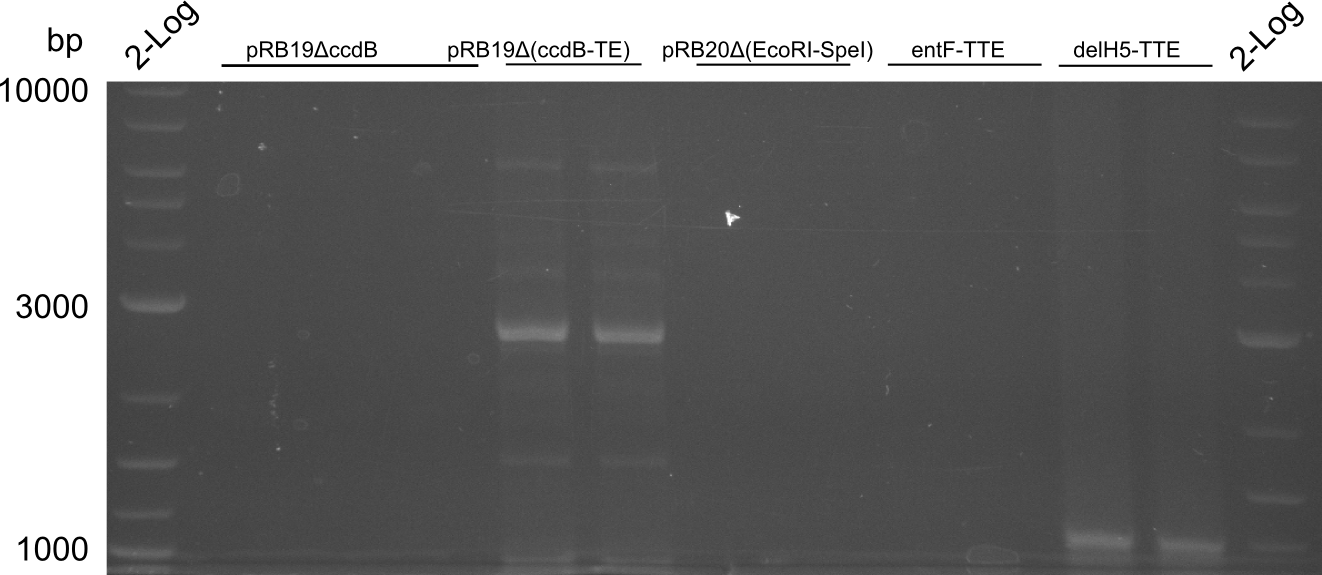
bpsA-TTE and tycC6-TTE worked, the other ones will be amplified using less stringent conditions
25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul
| TTE-Domains in BioRAD T100 | ||
|---|---|---|
| entF-TTE RB53/73 colony MG1655 | ||
| delH5-TTE RB61/75 colony D. aci SPH-1 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 120 |
| 10 | 98 | 1 |
| 62 ? 0.5 | 5 | |
| 72 | 20 | |
| 25 | 98 | 10 |
| 60 | 5 | |
| 72 | 20 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
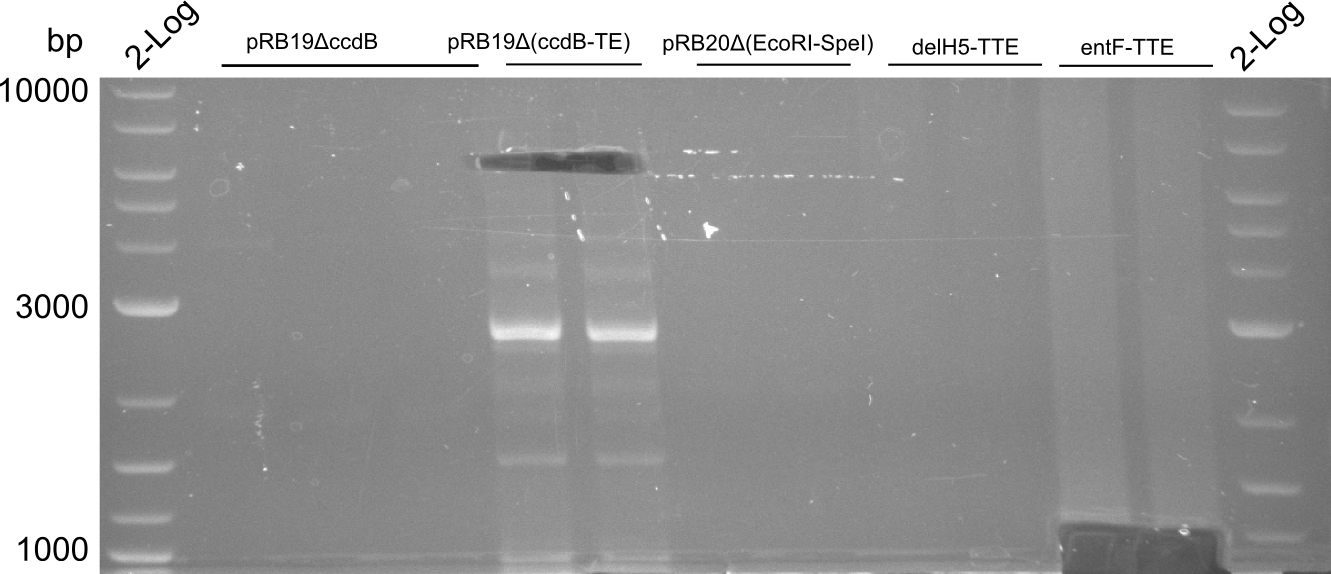
delH5-TTE worked, though with low yield. entF-TTE will be amplified in a third PCR.
25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul
| TTE-Domains in BioRAD T100 | ||
|---|---|---|
| entF-TTE RB53/73 colony MG1655 | ||
| cycles | temperature (°C) | time (s) |
| 1 | 98 | 120 |
| 10 | 98 | 1 |
| 60 ? 0.5 | 5 | |
| 72 | 20 | |
| 25 | 98 | 10 |
| 55 | 5 | |
| 72 | 20 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
entF-TTE didn't work. We will postpone this PCR.
T-Domain exchange
CPEC Assembly and Transformation
| Plasmid | Fragment 1 | Molarity [nM] | Volume in ul | Fragment 2 | Molarity [nM] | Volume in MM | CPEC Master Mix total [ul] |
|---|---|---|---|---|---|---|---|
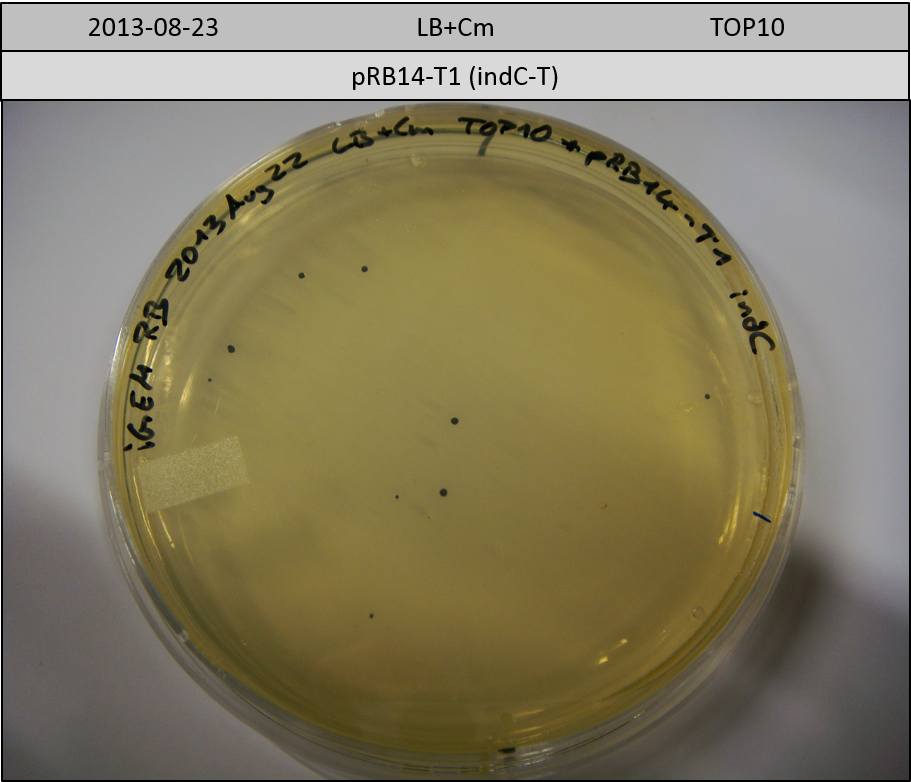
| pRB14-T1 | pRB14d(ccdB) | 3.1 | 2.7 | indC-T | 1457.6 | 0.3 | 6 |
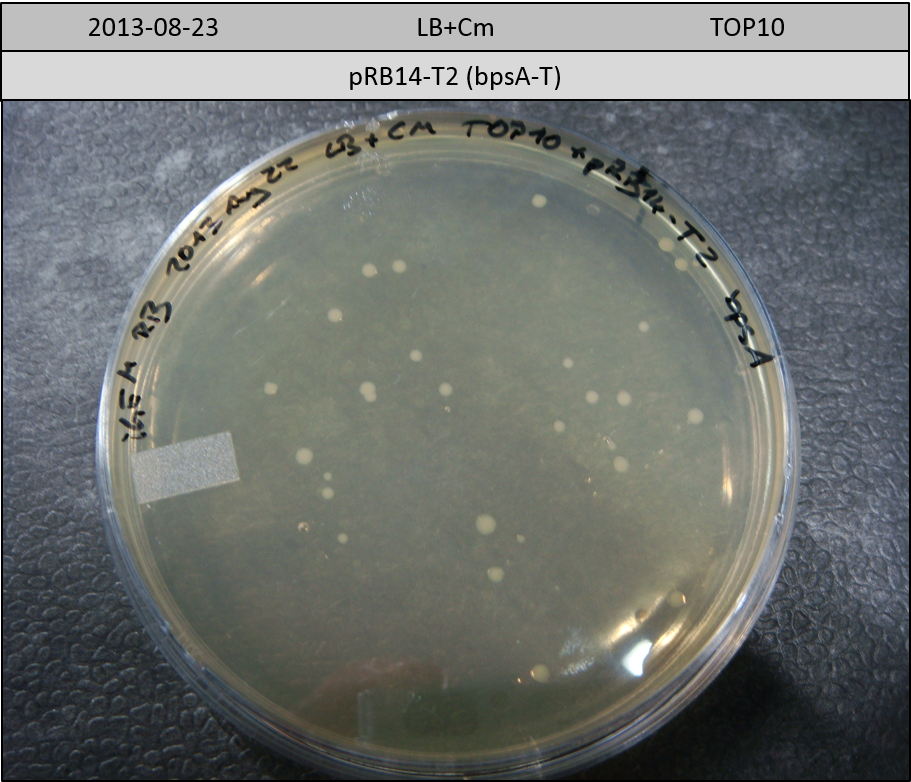
| pRB14-T2 | pRB14d(ccdB) | 3.1 | 2.7 | bpsA-T | 1506.1 | 0.3 | 6 |
| pRB14-T4 | pRB14d(ccdB) | 3.1 | 2.7 | tycA1-T | 1395.5 | 0.3 | 6 |
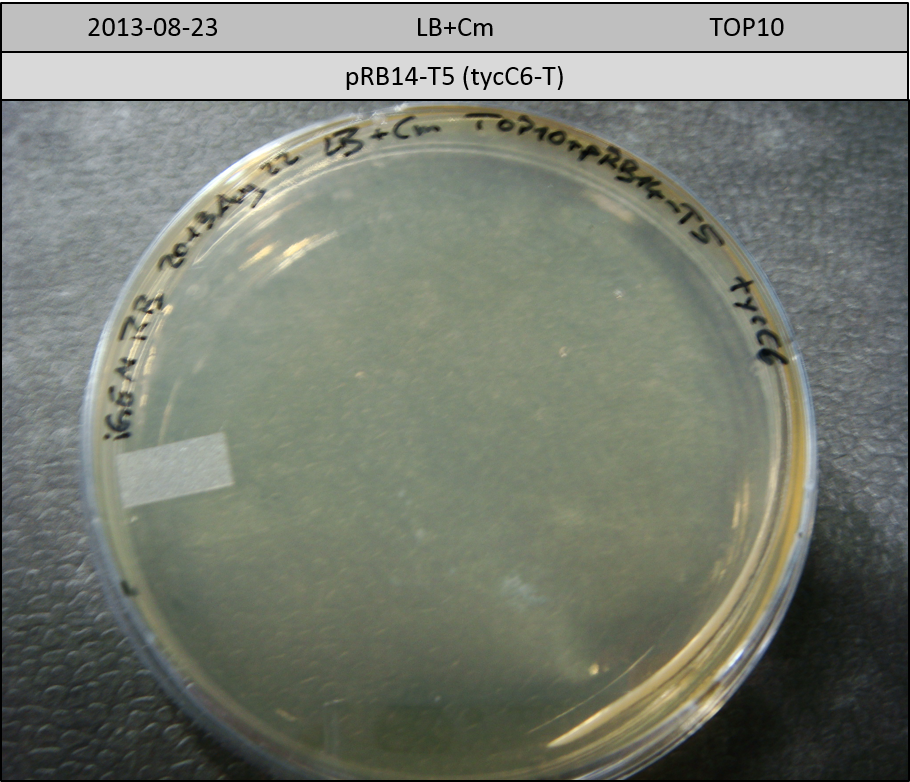
| pRB14-T5 | pRB14d(ccdB) | 3.1 | 2.7 | tycC6-T | 1504.5 | 0.3 | 6 |
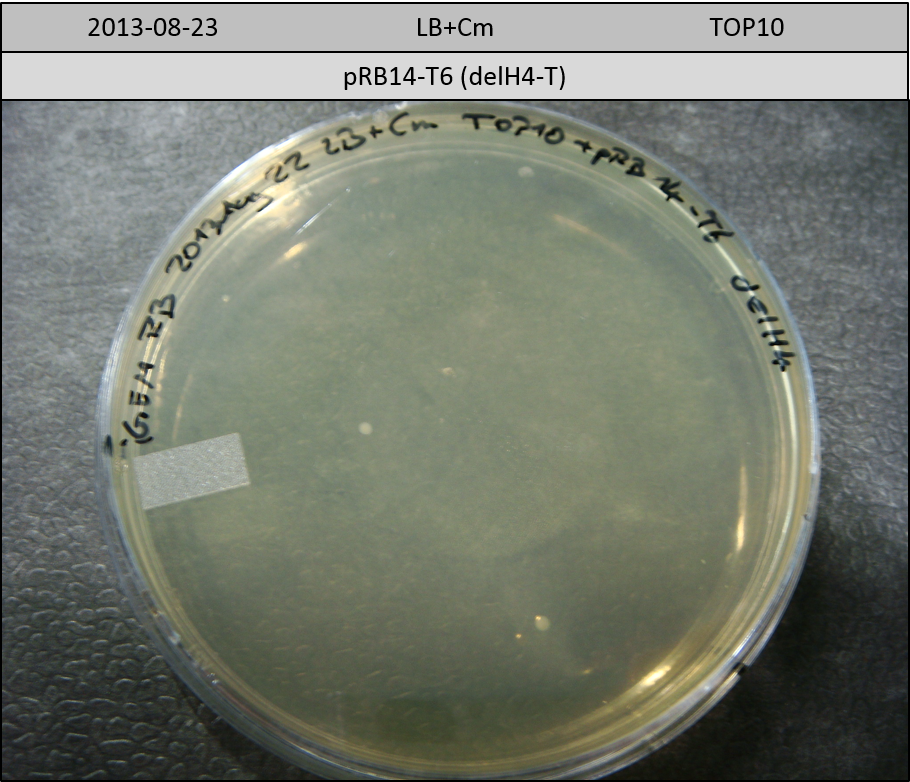
| pRB14-T6 | pRB14d(ccdB) | 3.1 | 2.7 | delH4-T | 1325.8 | 0.3 | 6 |
| pRB14-T7 | pRB14d(ccdB) | 3.1 | 2.7 | delH5-T | 1230.3 | 0.3 | 6 |
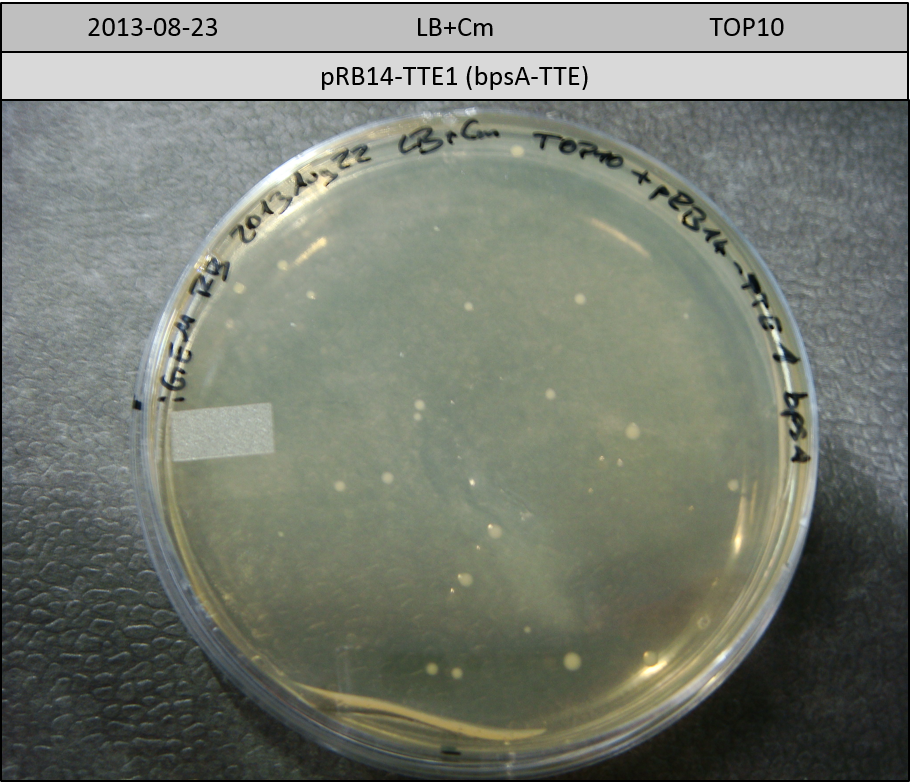
| pRB14-TTE1 | pRB14d(ccdB-TE) | 33.5 | 3 | bpsA-TTE | 140.8 | 2 | 10 |
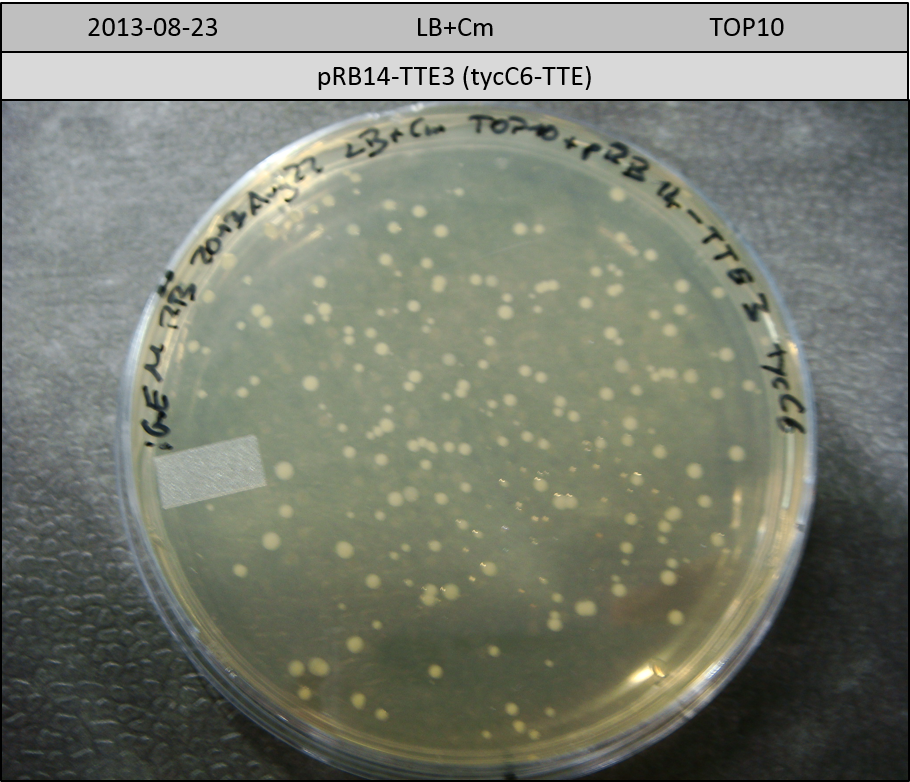
| pRB14-TTE3 | pRB14d(ccdB-TE) | 33.5 | 1.5 | tycC6-TTE | 31.8 | 3.5 | 10 |
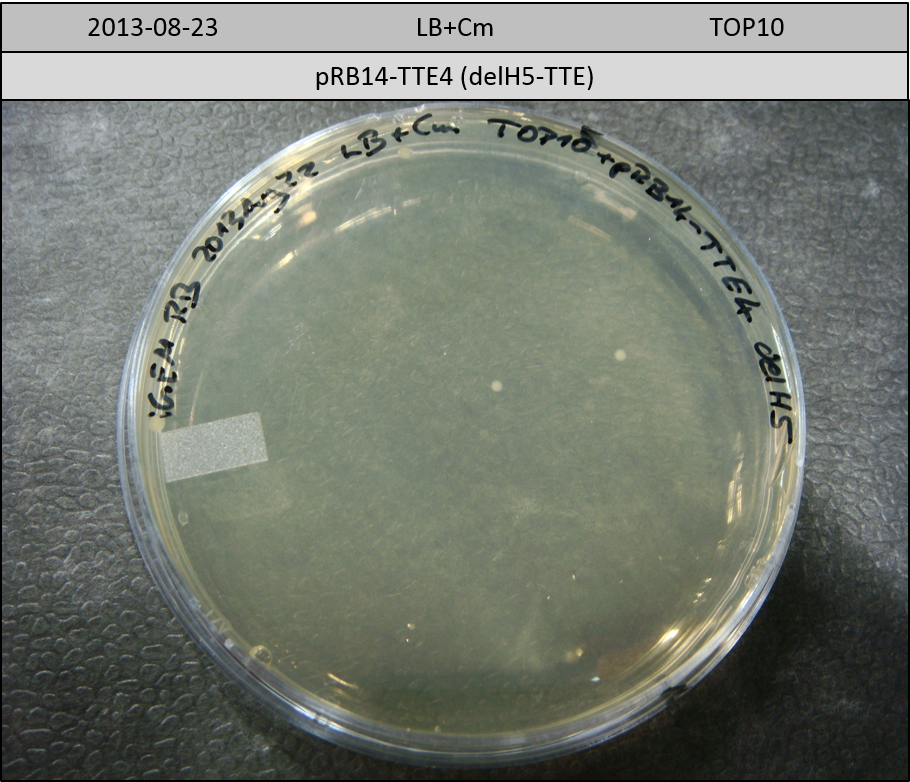
| pRB14-TTE4 | pRB14d(ccdB-TE) | 33.5 | 1.5 | delH5-TTE | 34.7 | 3.5 | 10 |
| BioRAD T100 | ||
| Cycles | Temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 10 |
| 8 | 98 | 1 |
| 53 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
Transformation in TOP10 according to standard protocol. Incubation for 30 hours at 37 °C and 10 hours
on the bench.
BioBricks
Fragment Preparation
25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul
| miniprep of pRB22 with RB68/69 | ||
| cycles | temperature (°C) | time (s) |
|---|---|---|
| 1 | 98 | 10 |
| 35 | 98 | 1 |
| 60 | 5 | |
| 72 | 100 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
PCR worked well. To make sure that we don't transform template plasmid pRB21, we digest the pRB22d(EcoRI-SpeI) fragment Gel Extraction together with pRB22d(SpeI-EcoRI) with DpnI for four hours at 37 °C.
| 37 °C; 4 hours | ||
| Component | Concentration | Volume [ul] |
|---|---|---|
| pRB22d(EcoRI-SpeI) | 32.3 nM | 11.0 |
| pRB22d(SpeI-EcoRi) | 839.4 nM | 2.4 |
| CutSmart buffer | 10x | 1.6 |
| NEB DpnI | 1 | |
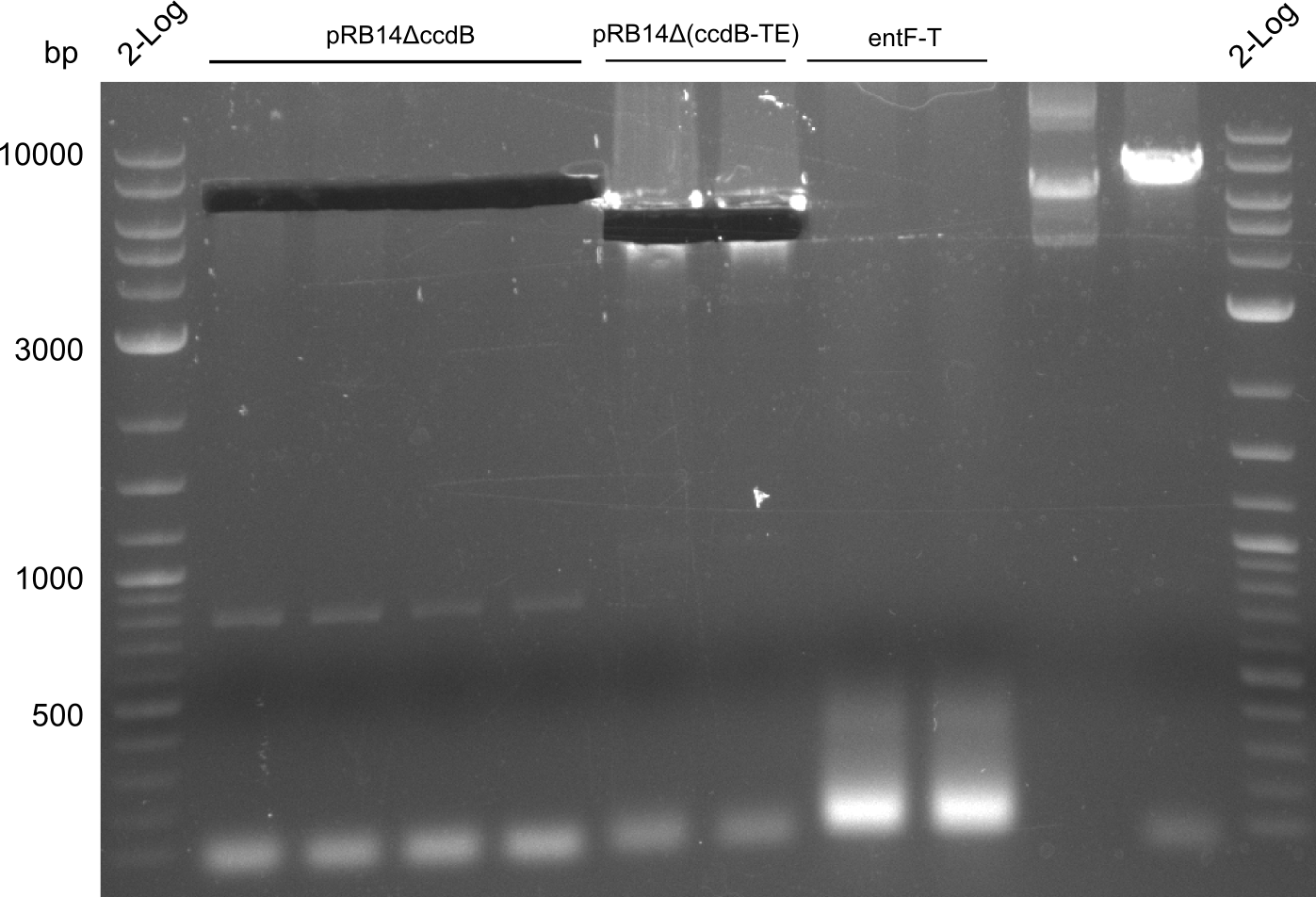
The Digest was purified using QiaGen Nucleotide Removal Kit. Unfortunately I dropped 2/3 onto the table... The final DNA concentration was 47.5 ng/ ul according to NanoVue. Standard CPEC assembly was performed in the old broken BioRAD cycler which didn't cool down to 53 °C but only to 58 °C for the annealing step. Hopefully it worked. TOP10 cells were transformed with 5 ul of CPEC master mix.
Transformation was successful. Ten colonies were screened using primers VF2 and KH6 in a 20 ul iTaq Master Mix according to standard protocol (50 °C).
Probes number x to x were purified using Qiaquick PCR purification kit and digested with NEB DpnI, EcoRI-HF and SpeI-HF over night to see whether pRB22 is really free of those restriction sites. The result was put on an AgaroseGel.
Lanes 1 to 4 show a slight smear beneath 200 bp. Lane 5 (probe 10) doesn't show that smear and the
specific band runs maybe 300 bp higher. So maybe the first four lanes contained restriction sites and
the last one didn't. We put probes 1 and 10 in 40 ml culture for midiprep. The midiprep will then be
digested as well so that we get certainity about the restriction sites. In parallel we did a colony PCR from probe 10 with KH3/4 using Phusion Flash HF to already have our
fragment for pRB23 for the T-Domain-exchange.
| miniprep of pRB23 with RB68/69 | ||
| cycles | temperature (°C) | time (s) |
|---|---|---|
| 1 | 98 | 120 |
| 10 | 98 | 1 |
| TD 62 | 5 | |
| 72 | 140 | |
| 25 | 98 | 1 |
| 57 | 5 | |
| 72 | 120 | |
| 1 | 72 | 300 |
| 1 | 12 | - |
 "
"