Team:Heidelberg/Templates/Indigoidine week18
From 2013.igem.org
| Line 1: | Line 1: | ||
| - | |||
| Line 33: | Line 32: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
{|class="wikitable" | {|class="wikitable" | ||
| Line 64: | Line 60: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
Gel Extraction with QIAquick gel extraction kit. | Gel Extraction with QIAquick gel extraction kit. | ||
| Line 83: | Line 76: | ||
Digest ran for 1 hour at 37 °C, was put on a agarose gel, gel extracted and eluted in 15 ul water. | Digest ran for 1 hour at 37 °C, was put on a agarose gel, gel extracted and eluted in 15 ul water. | ||
| - | |||
| - | |||
| - | |||
| - | |||
Concentration was measured using NanoVue: 30 ng/ ul. 10 ul were put together with 10 ul of Phusion | Concentration was measured using NanoVue: 30 ng/ ul. 10 ul were put together with 10 ul of Phusion | ||
| Line 93: | Line 82: | ||
OneShot and TOP10 cellc were transformed using different amounts of CPEC product. | OneShot and TOP10 cellc were transformed using different amounts of CPEC product. | ||
| - | |||
| - | |||
| - | |||
| - | |||
Ten colonies were screened using iTaq with KH9/VR and RB35/VR, respectively, to see whether the assembly worked and sfp was kicked out. | Ten colonies were screened using iTaq with KH9/VR and RB35/VR, respectively, to see whether the assembly worked and sfp was kicked out. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
The screening was positive, so probe 10 was kept for 10 ml liquid culture (TB+Cm). | The screening was positive, so probe 10 was kept for 10 ml liquid culture (TB+Cm). | ||
| Line 154: | Line 133: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
{|class="wikitable" | {|class="wikitable" | ||
| Line 179: | Line 155: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
Gel extraction and measurement using NanvoVue. The final table of T-domain concentration is as | Gel extraction and measurement using NanvoVue. The final table of T-domain concentration is as | ||
| Line 266: | Line 239: | ||
|- | |- | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
PCR was run with with a fragment of the Tyrocidine group with slightly suboptimal conditions. It will be repeated using otimal conditions. | PCR was run with with a fragment of the Tyrocidine group with slightly suboptimal conditions. It will be repeated using otimal conditions. | ||
| Line 300: | Line 269: | ||
|} | |} | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Gel extraction yielded 144.2 ng/ ul (Nanodrop) | Gel extraction yielded 144.2 ng/ ul (Nanodrop) | ||
| Line 325: | Line 291: | ||
Cells were plated on LB+Kanamycin and incubated at 37 °C for 24 hours. | Cells were plated on LB+Kanamycin and incubated at 37 °C for 24 hours. | ||
| - | + | ||
| - | + | ||
| - | + | ||
Five colonies of each plate were screened using iTaq polymerase and two primer combinations for each colony. This is VF2/PPTase_rv primer and PPTase_fw/VR primer. | Five colonies of each plate were screened using iTaq polymerase and two primer combinations for each colony. This is VF2/PPTase_rv primer and PPTase_fw/VR primer. | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Plasmids#pRB16|pRB16]] is obviously wrong. We checked the PCR product we used for the assembly and recognized that it's wrong, so we have to amplify it again from S. verticillus. DelC screenings show another small band, we have too check whether the primers bind elsewhere on the plasmid and run a negative control with those primers. | [[Plasmids#pRB16|pRB16]] is obviously wrong. We checked the PCR product we used for the assembly and recognized that it's wrong, so we have to amplify it again from S. verticillus. DelC screenings show another small band, we have too check whether the primers bind elsewhere on the plasmid and run a negative control with those primers. | ||
Latest revision as of 15:45, 21 October 2013
Contents |
Preparation for T-Domain exchange
Assembly of pRB19
PCR indC(ccdB) RB27/28 and pSB1C3 RB21/22 25 ul Phusion Flash HF MM 2x; 5 ul Primer 10 uM each; template according to table; water ad 50 ul
| RB27/46 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 5 | 98 | 1 |
| 55 | 5 | |
| 72 | 60 | |
| 30 | 98 | 1 |
| 72 | 60 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
| RB27/46 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 10 |
| 10 | 98 | 1 |
| td 55 | 5 | |
| 72 | 70 | |
| 25 | 98 | 1 |
| 60 | 5 | |
| 72 | 70 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
Gel Extraction with QIAquick gel extraction kit.
| Plasmid | Fragment 1 | Molarity [nM] | Volume in ul | Fragment 2 | Molarity [nM] | Volume in MM | DpnI
Master Mix total [ul] |
|---|---|---|---|---|---|---|---|
| pRB19 | indC(ccdB) from pRB14 | 14.6 | 14 | pSB1C3 RB21/22 | 105.3 | 3 | 20 |
Digest ran for 1 hour at 37 °C, was put on a agarose gel, gel extracted and eluted in 15 ul water.
Concentration was measured using NanoVue: 30 ng/ ul. 10 ul were put together with 10 ul of Phusion
Flash HF Master Mix for standard CPEC assembly. OneShot and TOP10 cellc were transformed using different amounts of CPEC product.
Ten colonies were screened using iTaq with KH9/VR and RB35/VR, respectively, to see whether the assembly worked and sfp was kicked out.
The screening was positive, so probe 10 was kept for 10 ml liquid culture (TB+Cm). Miniprep of 4 ml yielded 69.4 ng/ ul in 50 ul. Miniprep was used for PCR with primers KH3/4 to get the fragment pRB19 without ccdB for insertion of T-Domains via CPEC. First we tried two different polymerases at two different conditions each. The first is Phusion Flash
HF 2x Master Mix (ThermoScientific) and the other is Phusion HF 2x Master Mix (NEB).
| KH3/4 in BioRAD T100 with Phusion HF (NEB) | ||
|---|---|---|
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 30 |
| 30 | 98 | 15 |
| 57-60 | 15 | |
| 72 | 3.30 | |
| 1 | 72 | 10.00 |
| 1 | 12 | - |
| KH3/4 in BioRAD T100 with Phusion Flash HF (ThermoScientific) | ||
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 30 | 98 | 1 |
| 57-60 | 5 | |
| 72 | 2.00 | |
| 1 | 72 | 6.00 |
| 1 | 12 | - |
| KH5/6 in BioRAD T100 | ||
|---|---|---|
| cycles | temperature [°C] | time [s] |
| 1 | 98 | 30 |
| 45 | 98 | 5 |
| 60 | 15 | |
| 72 | 15 | |
| 1 | 72 | 60 |
| 1 | 12 | - |
Gel extraction and measurement using NanvoVue. The final table of T-domain concentration is as
follows:
| Fragment | Concentration [ng/ ul] | ~ Fragment size [bp] | Molarity [nM] |
|---|---|---|---|
| ccdB | 121.2 | 700 | 262.33 |
| indC-T | 192.4 | 200 | 1457.58 |
| bpsA-T | 198.9 | 200 | 1506.18 |
| entF-T | 64.5 | 200 | 488.6 |
| tycA1-T | 184.2 | 200 | 1395.45 |
| tycC6-T | 198.6 | 200 | 1504.55 |
| delH4-T | 175.0 | 200 | 1325.76 |
| delH5-T | 162.4 | 200 | 1230.30 |
| plu2642-T | 79.0 | 200 | 598.5 |
| plu2670-T | 31.5 | 200 | 238.6 |
| synT1 | 43 | 200 | 325.8 |
| synT2 | 37.5 | 200 | 284.1 |
| synT3 | 40.5 | 200 | 306.8 |
| synT4 | 36 | 200 | 272.7 |
| synT5 | 37.5 | 200 | 284.1 |
| synT6 | 37 | 200 | 280.3 |
| synT7 | 40.5 | 200 | 306.8 |
| bpsA-TTE | 92.9 | 1000 | 140.8 |
| entF-TTE | - | 1000 | - |
| tycC6-TTE | 21 | 1000 | 31.8 |
| delH5-T | 22.9 | 1000 | 34.7 |
Assembly of pRB15-18
pRB15-18 were unable to be sequenced with various primers. We now use pSB3K3 backbone from plate 3 of the 2013 spring distribution and assemble those PPTase-plasmids de novo.
| RB21/63 in BioRAD T100 | ||
|---|---|---|
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 14 | 98 | 1 |
| td 66 | 5 | |
| 72 | 60 | |
| 16 | 98 | 1 |
| 68 | 5 | |
| 72 | 60 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
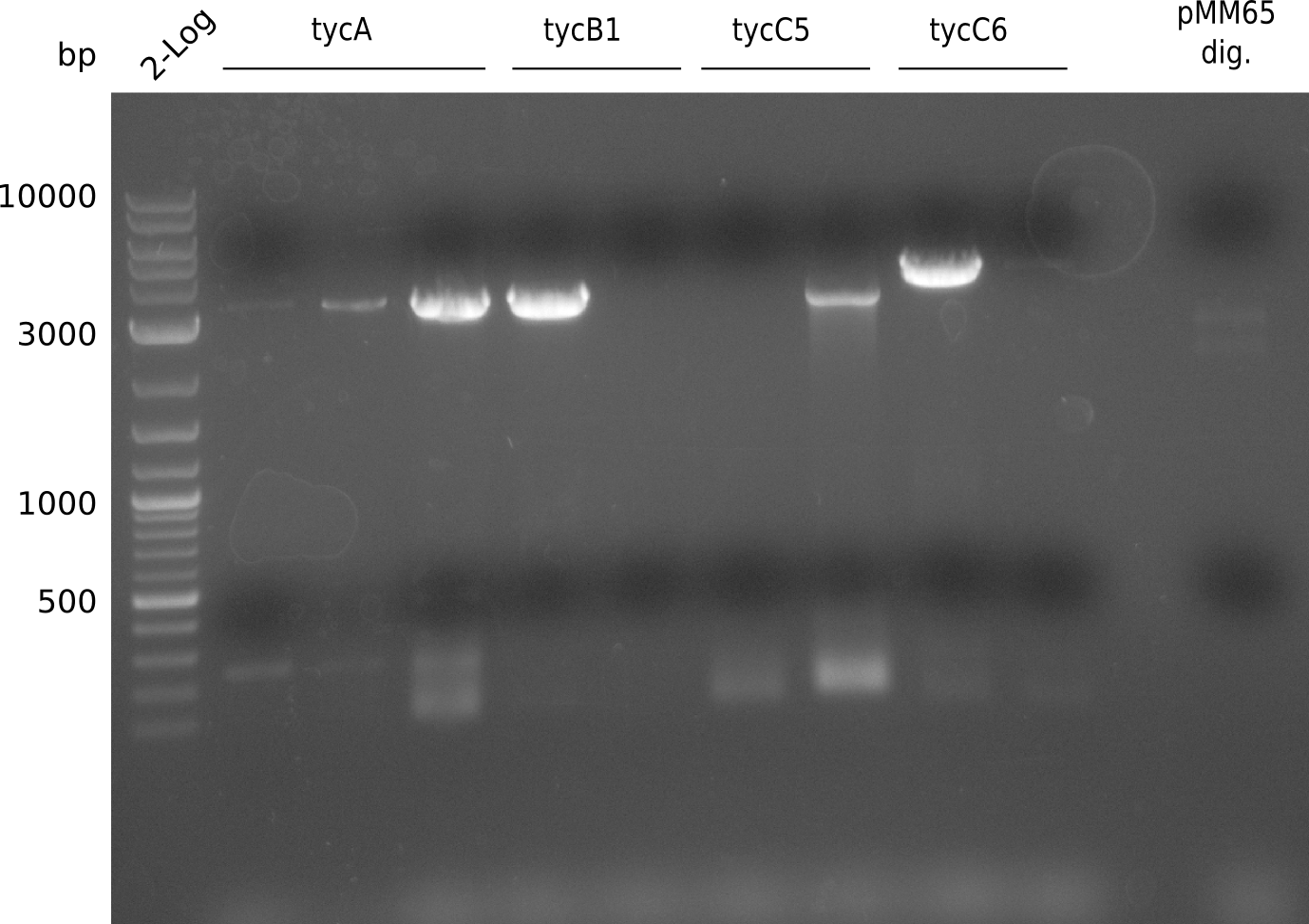
PCR was run with with a fragment of the Tyrocidine group with slightly suboptimal conditions. It will be repeated using otimal conditions.
| RB21/63 in BioRAD T100 | ||
|---|---|---|
| Cycles | Temperature [°C] | Time [s] |
| 1 | 98 | 10 |
| 12 | 98 | 1 |
| td 61 | 5 | |
| 72 | 50 | |
| 20 | 98 | 1 |
| 65 | 5 | |
| 72 | 50 | |
| 1 | 72 | 180 |
| 1 | 12 | - |
Gel extraction yielded 144.2 ng/ ul (Nanodrop)
CPEC was performed using pSB3K3 gel extraction and gel extractions made for assembly of pRB3-10. TOP10 cells were transformed using standard protocol.
| Plasmid | Fragment 1 | Molarity [nM] | Volume [ul] | Fragment 2 | Molarity [nM] | Volume [ul] | CPEC Master Mix total [ul] |
|---|---|---|---|---|---|---|---|
| pRB15 | pSB3K3(RB21/63) | 87.4 | 2 | sfp | 713.4 | 1 | 6 |
| pRB16 | pSB3K3(RB21/63) | 87.4 | 2 | svp | 90.7 | 4 | 12 |
| pRB17 | pSB3K3(RB21/63) | 87.4 | 2 | entD | 674.1 | 1 | 6 |
| pRB18 | pSB3K3(RB21/63) | 87.4 | 1 | delC | 118.1 | 2 | 6 |
Cells were plated on LB+Kanamycin and incubated at 37 °C for 24 hours.
Five colonies of each plate were screened using iTaq polymerase and two primer combinations for each colony. This is VF2/PPTase_rv primer and PPTase_fw/VR primer.
pRB16 is obviously wrong. We checked the PCR product we used for the assembly and recognized that it's wrong, so we have to amplify it again from S. verticillus. DelC screenings show another small band, we have too check whether the primers bind elsewhere on the plasmid and run a negative control with those primers.
Assembly of pKH4
pKH4 is a pRB3 derived version of an indigoidine synthetase expression plasmid w/o sfp as activating PPtases. So the two plasmid strategy can be conducted (at first) with this plasmid instead to rely on pRB21. Basically the idea is to use restriction sites before and after sfp to discard it and to religate with another sequence to yield a functional backbone.
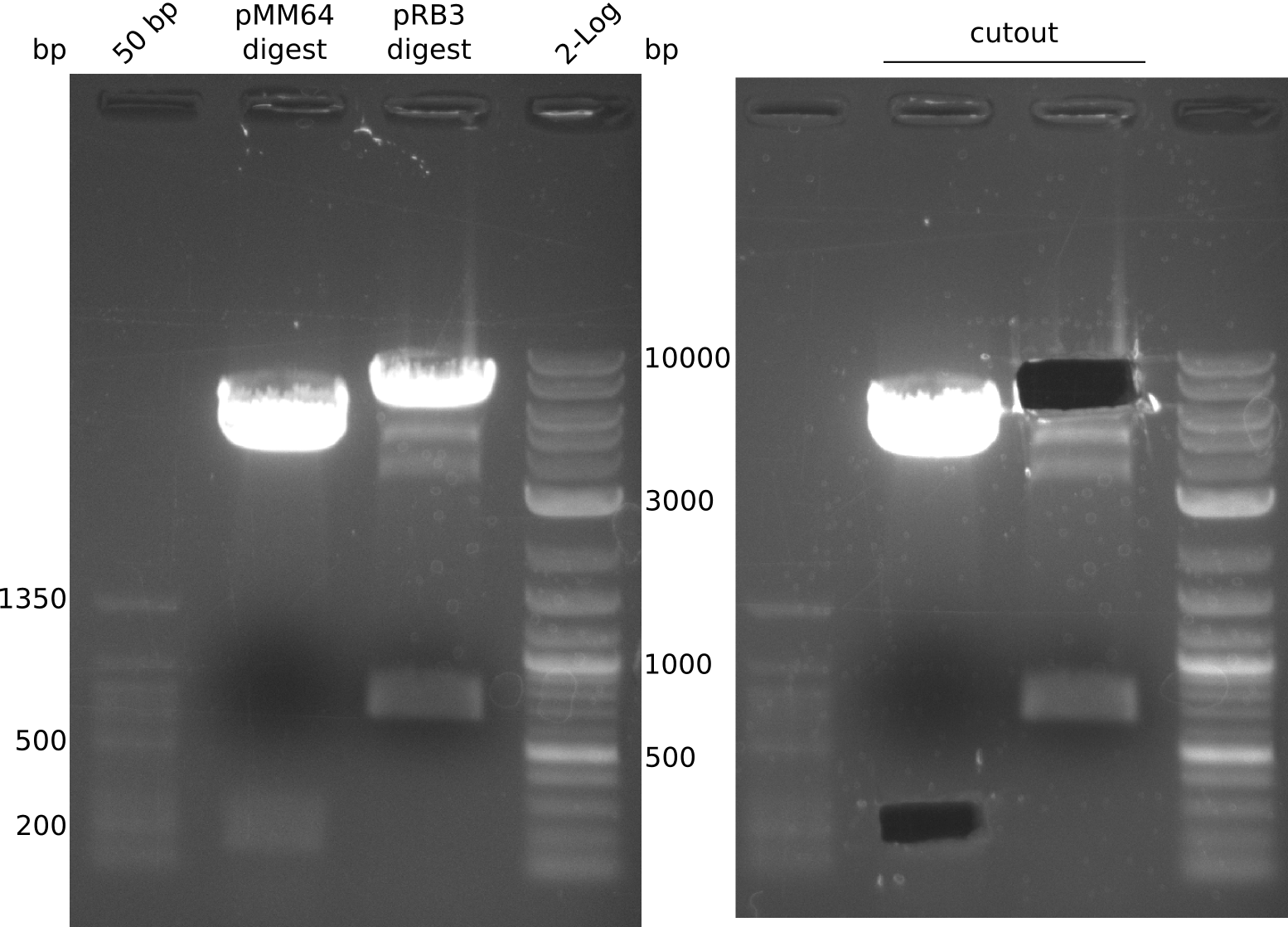
pRB3 has a BamHI before and a NheI cutting site after sfp. pMM64 and pMM65 both can be cut with a combination of BglII and XbaI to yield for example a 164 bp long fragment with compatible ends to the linearized pRB3. This fragment unfortunately contains a T7lac hybrid promoter which could interfere with our used primers, but still is worth a shot.
- digest mix volume: 30 µl (3 µl NEBuffer 3.1, 6.5 µl pMM65 (~481 ng), 0.3 µl (BglII, XbaI) each, 19.9 µl H2O)
- digestion gave expected bands at 2 kbp and 2.4 kbp but band at ~160 bp is missing. Reaction was carried out with not enough DNA.
- prepared ON for pMM64/65 MP as well as pRB3 MP
- the next day ON were extended to 2x 4 ml LB cultures which were then
prepped after several hours of incubation at 37 °C
- MP with cultures: DNA conc. measurement with NanoVue gave: 280
ng/µl (pMM64), 225 ng/µl (pMM65), 250 ng/µl (pRB3); analysis gel shows different concentration for pMM65 (~15 ng/µl ?)
- prepare digestion (20 µl), 37 °C, 1.5 h:
- pMM64: 2 µl NEBuffer 3.1, 17 µl pMM64, 0.5 µl (BglII, XbaI)
- pRB3: 2 µl NEBuffer CutSmart, 10 µl pRB3, 0.5 µl (BamHI, NheI), 7
µl H2O
- after gelextraction (elution in 20 µl) ligation was conducted with 2
µl 10x T4 ligase buffer, 1 µl T4 ligase, 2 µl pRB3 Dig_GE, 8 µl pMM64 Dig_GE and 7 µl H2O for 40 min at RT
- transformation in TOP10 with 1 µl and 5 µl of ligation mix
 "
"