Team:Heidelberg/Templates/MM week6 overview
From 2013.igem.org
Goal
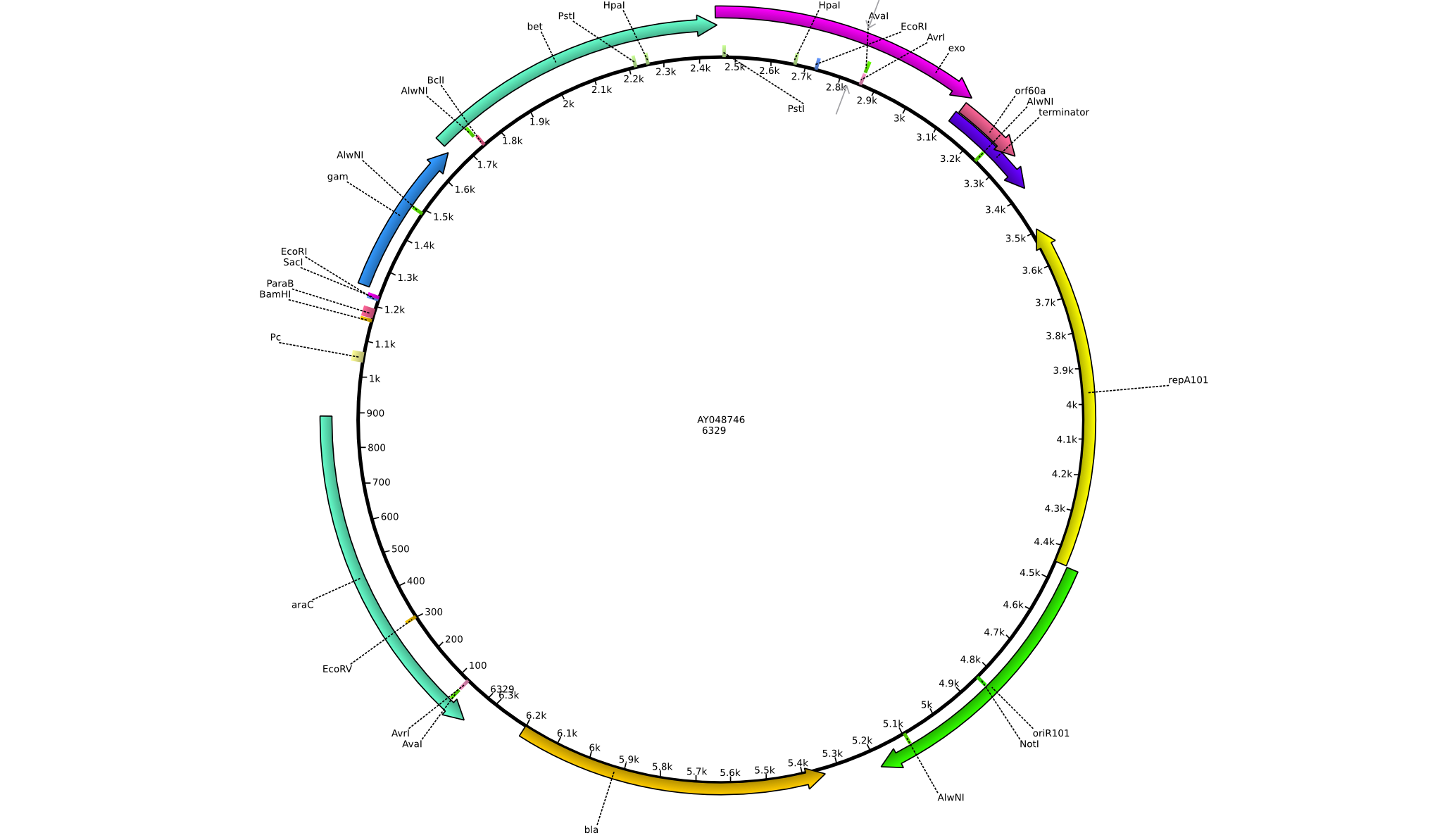
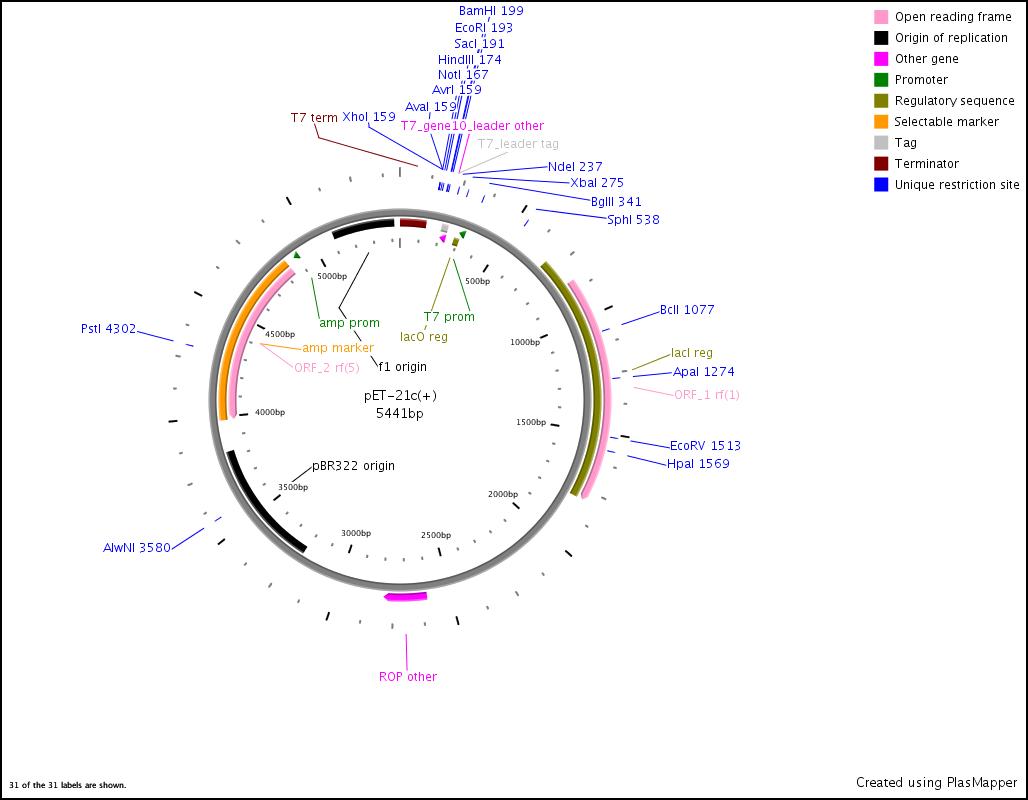
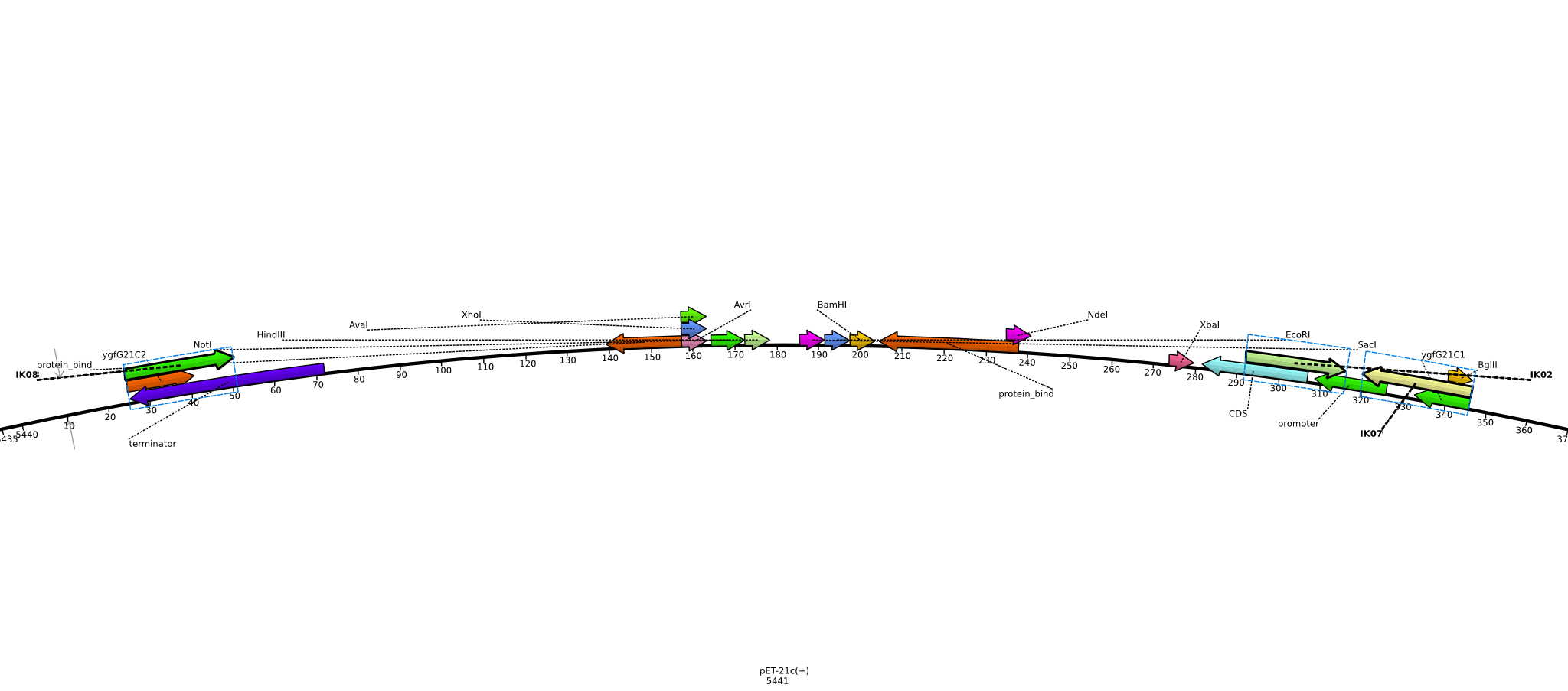
Integration of the methylmalonyl-CoA sythesis pathway into the BAP1 genome. This is a reproduction of a part of <bib id="pmid17959404"/>. For the integration, the λ-red protocol established by Datsenko and Wanner<bib id="pmid10829079"/> is used. pKD20/pKD46 plasmids contain the λ-red genes begind an arabinose inducible promoter, a temperature-sensitive origin, and an ampicillin selection marker. The part to be integrated is on the pLF03 plasmid, which is identical to pYW201<bib id="pmid17959404"/> in every way except for the selection marker in the insert (pYW201 has kanamycin resistance, pLF03 chloramphenicol). pLF03 contains an ampicillin marker that is expressed constitutively, chloramphenicol is only used to test for successful integration, as it is behind an IPTG-inducible T7 promoter (as are the other two genes, pccB and accA1). pLF03 is derived from pET-21c. The expected amplificate size for PCR of pLF03 is 4kb. pccB-accA will replace the ygfG gene in the BAP1 genome.
 "
"