Team:Heidelberg/Tyrocidine week18 interms
From 2013.igem.org
(Created page with " == Tyrocidine-Indigoidine-Fusion - Cloning & Validation == === Gibson Assembly === Gibson assemblies were prepared according to the following formulas: ====pPW03 (Phe-Ind)====...") |
|||
| Line 1: | Line 1: | ||
| - | + | ||
== Tyrocidine-Indigoidine-Fusion - Cloning & Validation == | == Tyrocidine-Indigoidine-Fusion - Cloning & Validation == | ||
=== Gibson Assembly === | === Gibson Assembly === | ||
| Line 60: | Line 60: | ||
<gallery> | <gallery> | ||
| - | File: | + | File:Heidelberg_BAP_12_Eco.png | pPW03 and pPW04 digested with EcoRI, samples A, B, C & D for pPW03 and B, C & D for pPW04 show the expected bands. |
| - | File: | + | File:Heidelberg_BAP_3_Eco__BAP_TetI_EcoPst.png | pPW05 digested with EcoRI, all samples show the expected bands. The six lanes on the left are neglectable. |
</gallery> | </gallery> | ||
| Line 76: | Line 76: | ||
* pPW05 (Val-Ind): 5858 and 3105 | * pPW05 (Val-Ind): 5858 and 3105 | ||
| - | [[File: | + | [[File:Heidelberg_BAP_123_ECORI.png|300px]] |
As these results were not as clear as for the sequenced samples, we did not continue work on the direct BAP-transformants. | As these results were not as clear as for the sequenced samples, we did not continue work on the direct BAP-transformants. | ||
| Line 86: | Line 86: | ||
==== Results ==== | ==== Results ==== | ||
<gallery> | <gallery> | ||
| - | File: | + | File:Heidelberg_COLPCR_03_04_05_BAP_Tyc.png|Screening for the Tyc-part in blue (B1, B2, B3) and white (W1, W2) colonies from different plates with cells that were transformed with pPW03, pPW04 or pPW05 |
| - | File: | + | File:Heidelberg_COLPCR_03_04_05_BAP_Ind.png|Screening for the Tyc-part in blue (B1, B2, B3) and white (W1, W2) colonies from different plates with cells that were transformed with pPW03, pPW04 or pPW05 (left of black line) and screening for the Ind-part (right of black line) |
</gallery> | </gallery> | ||
| Line 105: | Line 105: | ||
==== Results ==== | ==== Results ==== | ||
<gallery> | <gallery> | ||
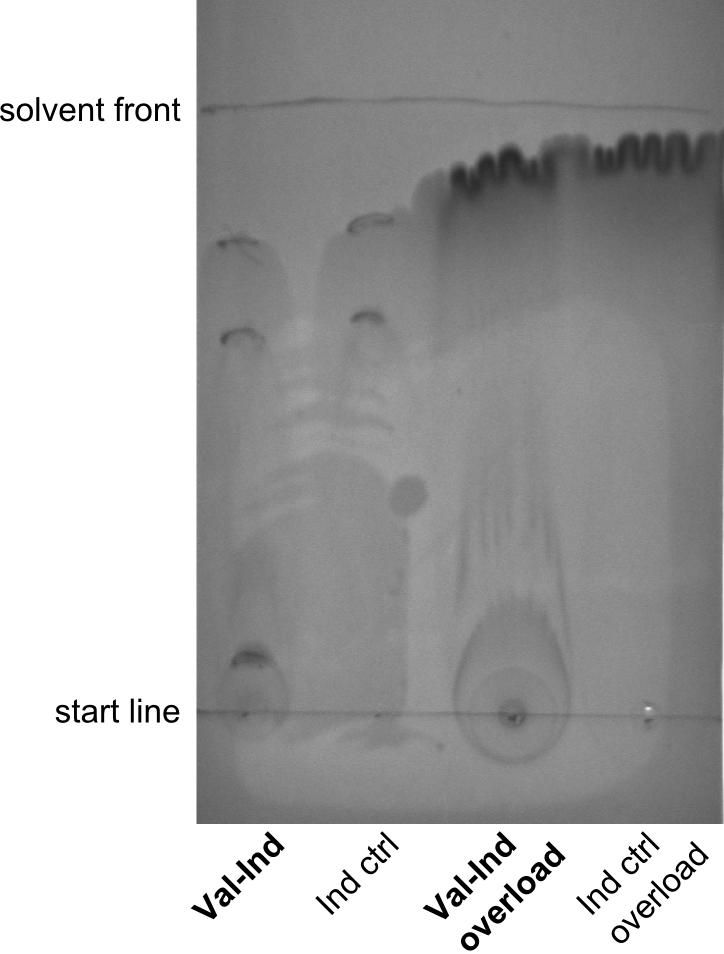
| - | File: | + | File:Heidelberg_TLC_1.png|first TLC - one biological replicate for Val-Ind and one biological replicate for Indigoidine, including the crude supernatants and methanol-washes were used. |
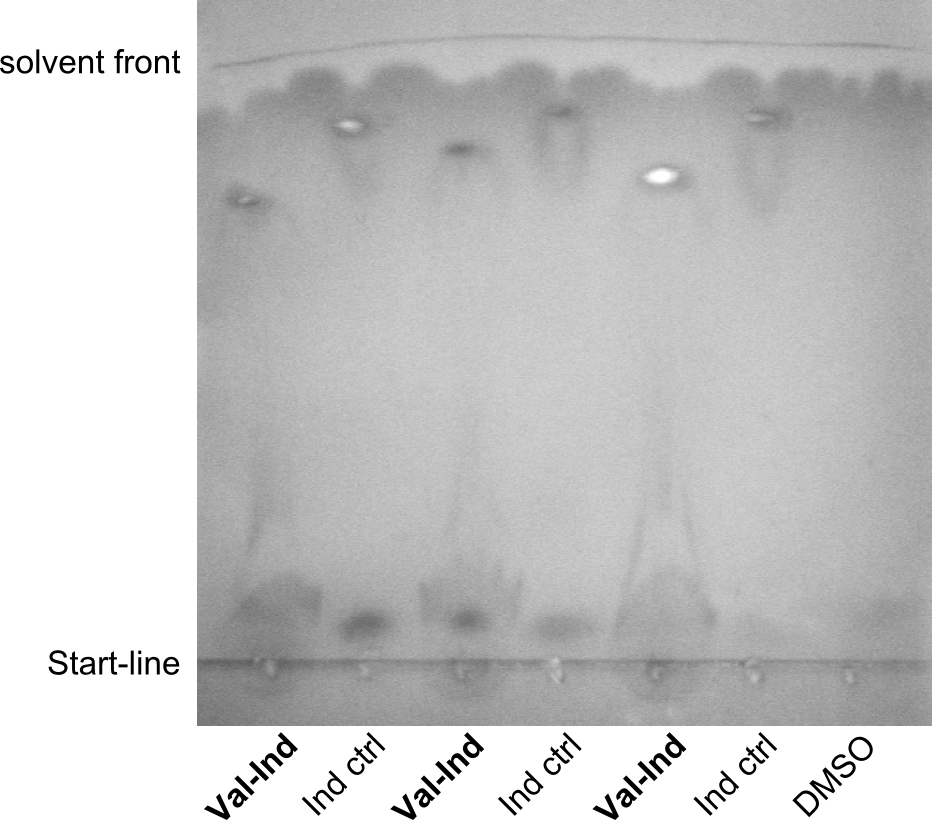
| - | File: | + | File:Heidelberg_TLC_2.png|second TLC |
| - | File: | + | File:Heidelberg_TLC_3.png|third TLC |
| - | File: | + | File:Heidelberg_TLC_4.png|fourth TLC |
| - | File: | + | File:Heidelberg_TLC_5.png|fifth TLC - showing the running behavior for three biological replicates of Val-Ind and four technical replicates of Indigoidine. Samples were applied alternatingly in order to exclude the possibility of different running behavior due to placement on the TLC. |
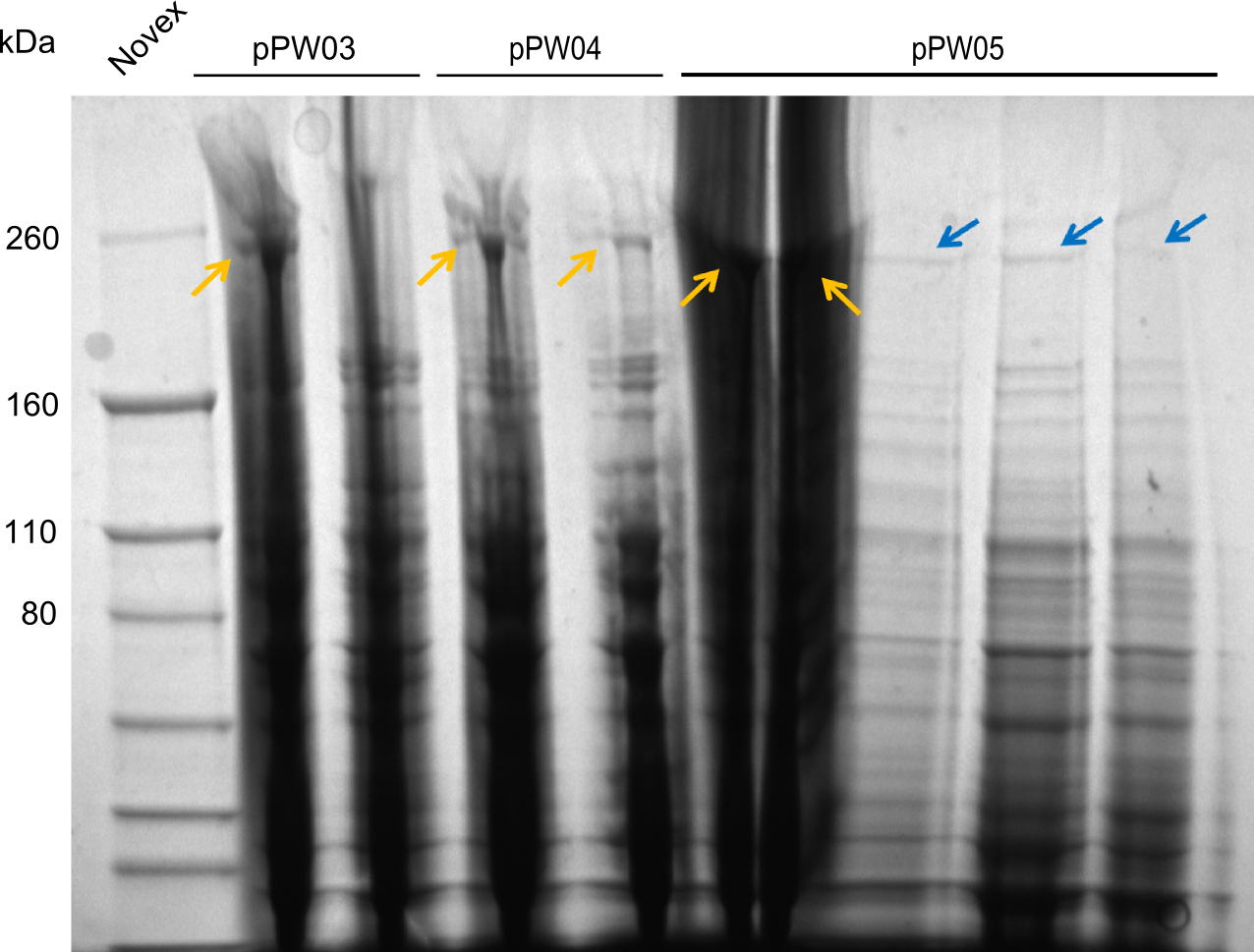
| - | File: | + | File:Heidelberg_SDS_3rd.png|SDS-PAGE, stained with Coomassie. The bands for the expected Val-Ind-NRPS are visible as indicated by the blue arrows. The golden arrows mark highly probable bands for the other NRPSs that are however qualitatively not 100% clear due to e.g. smear. |
</gallery> | </gallery> | ||
Latest revision as of 16:53, 4 October 2013
Contents |
Tyrocidine-Indigoidine-Fusion - Cloning & Validation
Gibson Assembly
Gibson assemblies were prepared according to the following formulas:
pPW03 (Phe-Ind)
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| A | 12 | 0.89 |
| B | 8 | 4.29 |
| C | 6 | 2.86 |
| D | 35 | 1.96 |
pPW04 (Asn-Ind)
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| D | 35 | 3.87 |
| E | 20 | 1.06 |
| F | 20 | 5.08 |
pPW05 (Val-Ind)
| fragment | concentration [ng/µl] | volume for gibson assembly [µL] |
|---|---|---|
| D | 35 | 3.45 |
| G | 15 | 1.26 |
| H | 16 | 3.78 |
| I | 20 | 1.51 |
Transformation
The constructs prepared by gibson assembly were chemically transformed (heat shock) into competent DH10ß.
Restriction Digest
Procedure
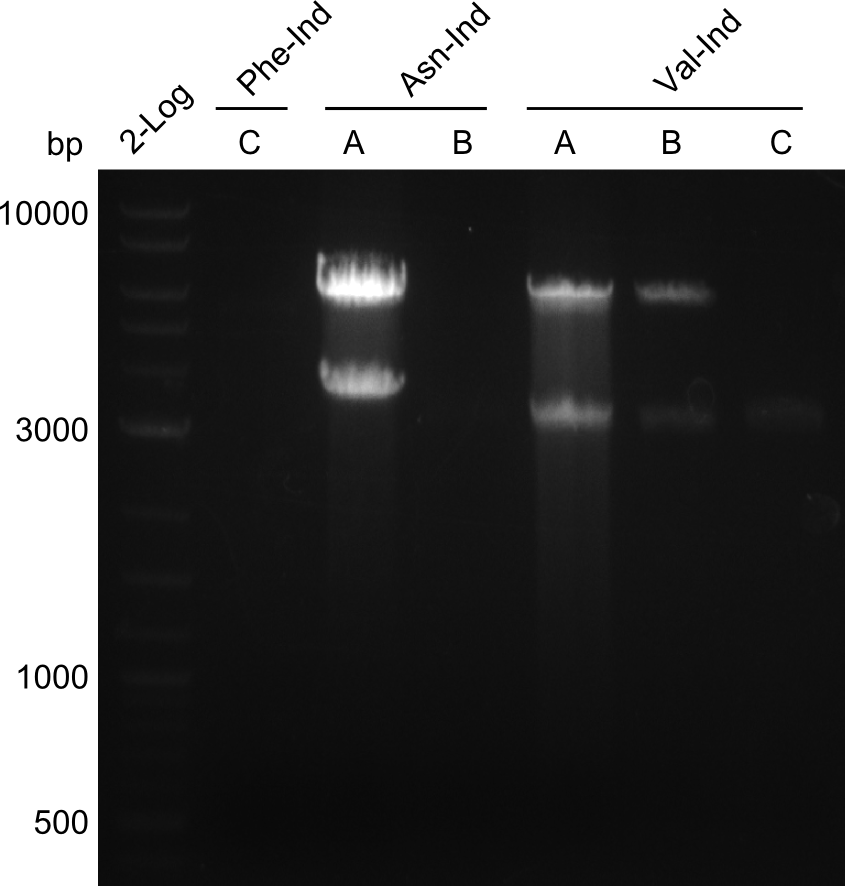
DH10ß - colonies were picked and grown overnight in LB with Chloramphenicol. After a miniprep, a restriction digest with EcoRI was performed, the expected bands were:
- pPW03 (Phe-Ind): 5858 and 3183
- pPW04 (Asn-Ind): 5858 and 3582
- pPW05 (Val-Ind): 5858 and 3105
Results
All samples showed the expected bands.
Discussion
Clones pPW03 A, B, C & D; pPW04 B, C & D; pPW05 A, B, C, D, E & F show the expected bands, hence two of each clone were sent to sequencing.
Sequencing & Transformation into BAP-I
Vectors were sequenced with the Primers used for the assembly. All sequences turned out to be positive, hence the minipreps that were sent to sequencing, were used for chemical transformation into BAP-I-cells accoding to the standard protocol. For each transformation 10µl and the rest were plated seperately. After less then two days, blue colonies occured on all plates, hence colony-PCR were performed on 5 colonies of each plate (3 blue ones and 2 white ones).
In parallel, BAP-I-cells were transformed with the remaining Gibson-Mix for pPW03, pPW04 and pPW05. Cells were picked and grown in LB with Chloramphenicol overnight. After miniprep, restricion digest with EcoRI was performed in 20 µl volume. Again, the expected bands were:
- pPW03 (Phe-Ind): 5858 and 3183
- pPW04 (Asn-Ind): 5858 and 3582
- pPW05 (Val-Ind): 5858 and 3105
As these results were not as clear as for the sequenced samples, we did not continue work on the direct BAP-transformants.
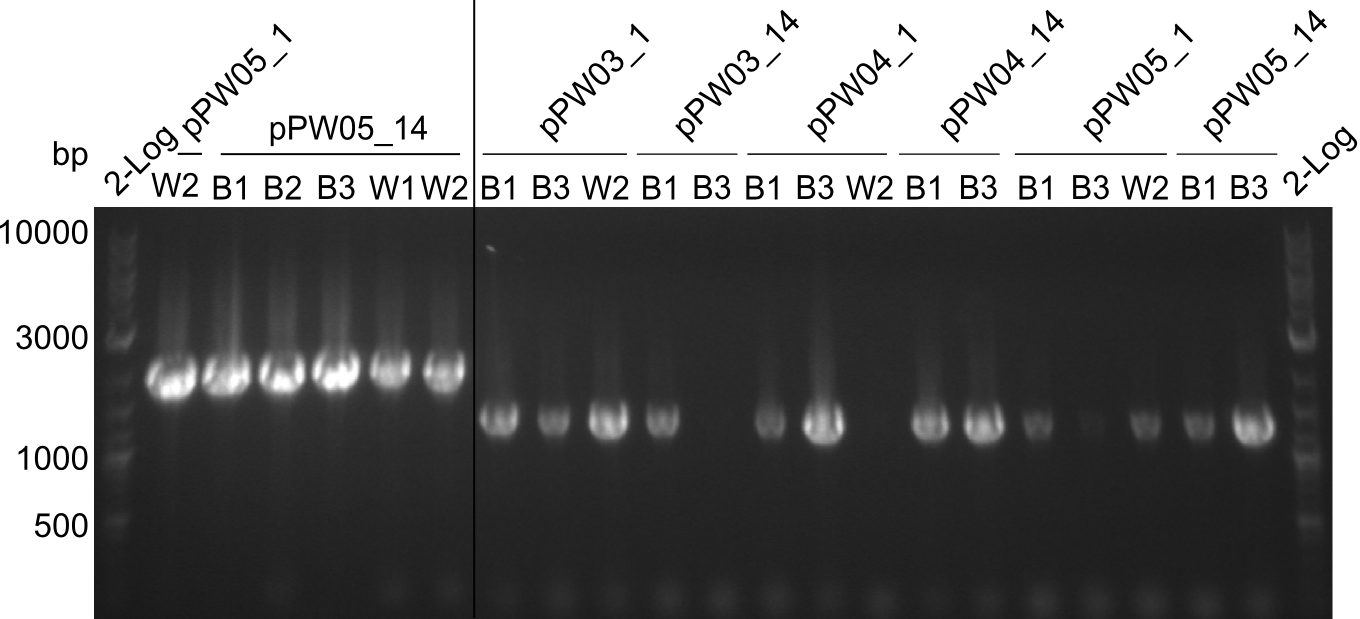
Screening PCR for positive BAP-I-colonies
Procedure
In order to screen for positive BAP-I-colonies, blue and white colonies were picked and used in colony-PCR.
Results
Almost all colonies were positive for both modules, except the colonies that were transformed with pPW03 (Phe-Ind). For these constructs, the bands that show up do not match the expected bands:
- expected: xxxx
- obtained: yyyy
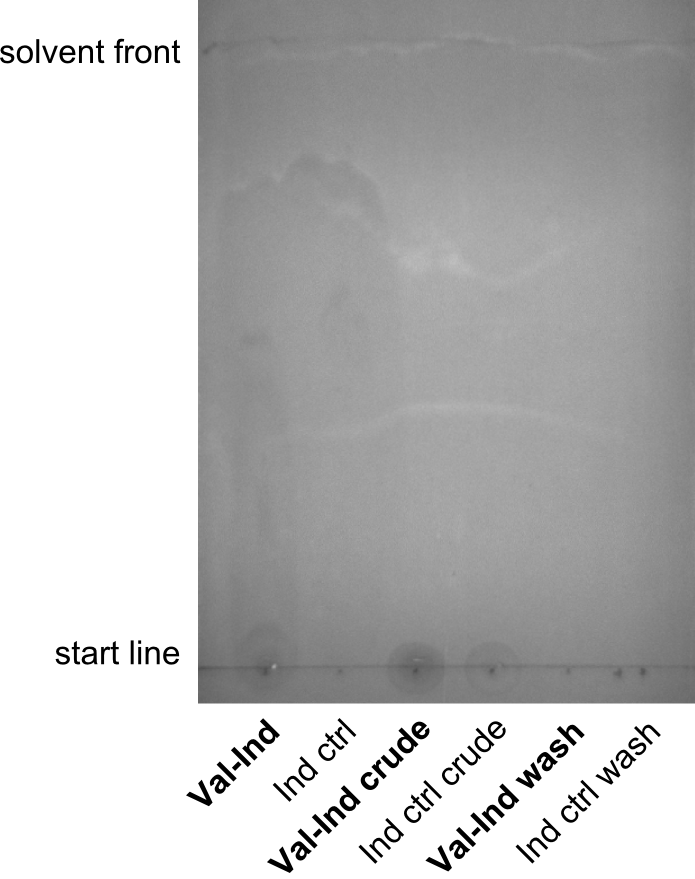
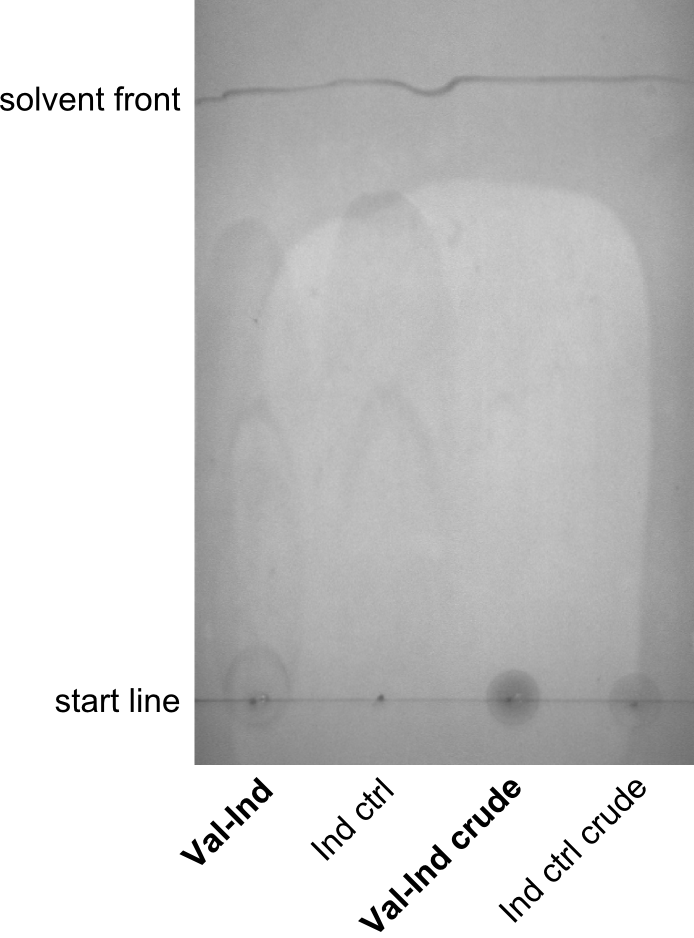
Induction, SDS-PAGE & TLC
Procedure
Cells were grown at 37°C and induced at OD600 = ~0.5 with 1mM IPTG. Cultures that were transformed with pPW05 turned blue after about 30 to 60 minutes. Cells were kept at 30°C overnight and the synthetized NRP was extracted and purified from samples. Herefore, 1ml culture was centrifuged at 14,000 rpm for 15 minutes, supernatant was kept for further analysis. The pellet was washed in 1ml Methanol and centrifuged at 14,000 rpm for 15 minutes. Supernatant was kept for further analysis and pellet was resuspended in 500µl DMSO. The crude supernatant, the methanol-wash and the purified NRP were used for TLC. As a control, purified indigoidine (treated with the same protocol) and its washes were used.
As solvent, a mixture of 95% Dichloromethane and 5% Methanol was used. However, all samples were running at the solvent front, hence a less polar solvent, i.e. 100% Dichloromethane was used, and qualitatively better results were obtained.
For the SDS-PAGE, remaining culture was treated as described previously and prepared for SDS-PAGE. Not only pPW05-cells, but also pPW03 and pPW04-positive cells were tested, in order to check for proper expression of the NRPSs.
Results
Discussion
Especially the last TLC-picture nicely shows that the indigoidine-control has a significantly and reproducibly different sunning behavior on the TLC. Different biological replicates (different clones) were used for Valin-Indigoidine and in all cases, it is running lower than the control - as expected.
As far as the SDS-PAGE is concerned, the expected protein sizes are:
- Phe-Ind-NRPS: 247 kDa
- Asn-Ind-NRPS: 261 kDa
- Val-Ind-NRPS: 242 kDa
- Note: the indigoidine-synthetase alone: 145 kDa
As indicated by the blue arrows, the Val-Ind-NRPS is clearly present in all of the tested samples. For the other two NRPSs, there are bands at the expected height, as indicated by the golden arrows, these are however qualitatively not as good as the ones for the Val-Ind-NRPS due to smear or low expression.
 "
"