Team:Hong Kong CUHK/backgroundVS
From 2013.igem.org
(Difference between revisions)
(Created page with "<html xmlns="http://www.w3.org/1999/xhtml"> <head> <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> <title>iGEM CUHK</title> <link rel="stylesheet" type="te...") |
Janeytling (Talk | contribs) |
||
| (8 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
<style type="text/css"> | <style type="text/css"> | ||
<!-- | <!-- | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
body{background-color:#660099} | body{background-color:#660099} | ||
#globalWrapper { | #globalWrapper { | ||
| Line 54: | Line 49: | ||
#content | #content | ||
{width: 1000px;border:none;background-color:#660099;padding:0px;margin-top:0px;line-height: 1.5em;color: black;} | {width: 1000px;border:none;background-color:#660099;padding:0px;margin-top:0px;line-height: 1.5em;color: black;} | ||
| - | #footnote { | + | #footnote {background-color:#FFFF93; |
position:absolute; | position:absolute; | ||
left:0; | left:0; | ||
| Line 76: | Line 71: | ||
#containerX2{ | #containerX2{ | ||
position:relative; | position:relative; | ||
| - | width: | + | width:1100px; |
| - | height: | + | height:1900px; |
| + | z-index:1; | ||
| + | background-color: #FFF; | ||
| + | } | ||
| + | #containerX4{ | ||
| + | position:relative; | ||
| + | width:1025px; | ||
| + | height:1900px; | ||
z-index:1; | z-index:1; | ||
background-color: #FFF; | background-color: #FFF; | ||
| Line 103: | Line 105: | ||
<div id="buttons"> | <div id="buttons"> | ||
| - | <div class="menubar" id="homeUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK"><img src="https://static.igem.org/mediawiki/2013/e/ee/Bpurple_home.png" width=" | + | <div class="menubar" id="homeUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK"><img src="https://static.igem.org/mediawiki/2013/e/ee/Bpurple_home.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="teamUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/team"><img src="https://static.igem.org/mediawiki/2013/d/d8/Bpurple_team.png" width=" | + | <div class="menubar" id="teamUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/team"><img src="https://static.igem.org/mediawiki/2013/d/d8/Bpurple_team.png" width="110" height="71" /></a></div> |
| - | <div class="menubar" id="projectUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/project"><img src="https://static.igem.org/mediawiki/2013/2/2d/Bpurple_project.png" width=" | + | <div class="menubar" id="projectUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/project"><img src="https://static.igem.org/mediawiki/2013/2/2d/Bpurple_project.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="biobrickUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/biobrick"><img src="https://static.igem.org/mediawiki/2013/b/b6/Bpurple_biobrick.png" width=" | + | <div class="menubar" id="biobrickUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/biobrick"><img src="https://static.igem.org/mediawiki/2013/b/b6/Bpurple_biobrick.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="hpUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/humanpractice"><img src="https://static.igem.org/mediawiki/2013/9/95/Bpurple_human_practice.png" width=" | + | <div class="menubar" id="hpUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/humanpractice"><img src="https://static.igem.org/mediawiki/2013/9/95/Bpurple_human_practice.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="modelUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/modelling"><img src="https://static.igem.org/mediawiki/2013/3/3f/Bpurple_modelling.png" width=" | + | <div class="menubar" id="modelUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/modelling"><img src="https://static.igem.org/mediawiki/2013/3/3f/Bpurple_modelling.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="safetyUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/safety"><img src="https://static.igem.org/mediawiki/2013/1/1e/Bpurple_safety.png" width=" | + | <div class="menubar" id="safetyUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/safety"><img src="https://static.igem.org/mediawiki/2013/1/1e/Bpurple_safety.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="docUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/documentation"><img src="https://static.igem.org/mediawiki/2013/1/12/Bpurple_documentation.png" width=" | + | <div class="menubar" id="docUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/documentation"><img src="https://static.igem.org/mediawiki/2013/1/12/Bpurple_documentation.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="galleryUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/gallery"><img src="https://static.igem.org/mediawiki/2013/e/ed/Bpurple_gallery.png" width=" | + | <div class="menubar" id="galleryUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/gallery"><img src="https://static.igem.org/mediawiki/2013/e/ed/Bpurple_gallery.png" width="110" height="71"/></a></div> |
| - | <div class="menubar" id="ackUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/acknowledgement"><img src="https://static.igem.org/mediawiki/2013/4/4b/Bpurple_ack.png" width=" | + | <div class="menubar" id="ackUP"><a href="https://2013.igem.org/Team:Hong_Kong_CUHK/acknowledgement"><img src="https://static.igem.org/mediawiki/2013/4/4b/Bpurple_ack.png" width="110" height="71"/></a></div> |
</div> | </div> | ||
| Line 125: | Line 127: | ||
<div id="middle"> | <div id="middle"> | ||
<div id="containerX2" style="margin: 0px auto;"> | <div id="containerX2" style="margin: 0px auto;"> | ||
| + | <div id="containerX4" style="margin: 0px auto;"> | ||
<div id="heading" style="margin: 0px auto;"> | <div id="heading" style="margin: 0px auto;"> | ||
<h2>Voltage Switch</h2> | <h2>Voltage Switch</h2> | ||
</div> | </div> | ||
<div id="partable"> | <div id="partable"> | ||
| - | + | <p2> | |
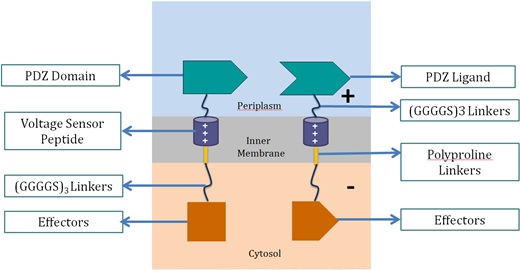
| - | + | <p>Voltage Switch is a response system triggered by a change of potential across the bacterial inner membrane. Components of this system include the transmembrane voltage sensor peptide originated from potassium ion channels, polyproline linker in the inner membrane, glycine serine linker, PDZ domain and PDZ ligand in the periplasm, and glycine serine linker and effectors in the cytosol.</p> | |
| - | + | <h3>1. Components of Voltage Switch System</h3><p align="center"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2013/8/84/Vs1.png" alt="2" width="521" height="270"> <br> | |
| - | + | </p> | |
| - | + | <p><stong> | |
| - | + | <strong>Voltage Sensor Peptide</strong></strong><br> | |
| - | + | Voltage sensor peptide, originated from the archaea <em>Aeropyrum pernix</em>, is a protein domain found in a voltage-dependent potassium ion channel. Previous studies suggested that the domain could translocate within the membrane upon membrane potential change [10, 11]. This helix peptide received its voltage-sensing ability from its four positively-charged arginine residues, and high hydrophobicity from its numerous leucine residues.</p> | |
| - | + | <p><strong>PDZ Domain and PDZ Ligand</strong><br> | |
| - | + | This is a dimer found in <em>Mus musculus</em>, and were used by the 2012 SJTU iGEM team as a membrane scaffolding tool. The PDZ dimer provides the necessary linkage of the two fusion proteins in the voltage switch system.</p> | |
| - | + | <p><strong>Polyproline Linker</strong><br> | |
| - | + | Polyproline linker is a pure proline rigid rod. Its rigidity is due to the limited rotation of peptide bonds in proline, a ring-shaped amino acid. Also, proline does not contain any hydrophilic functional group, which makes the linker hydrophobic and thus able to stay within the inner membrane.</p> | |
| - | + | <p><strong>Glycine Serine Linker</strong><br> | |
| - | + | This is a hydrophilic flexible linker which links PDZ dimer with voltage sensor peptides. It also links the polyproline linkers in the membrane with the effectors in the cytosol.</p> | |
| - | + | <p><strong>Effectors</strong><br> | |
| - | + | These effectors can be changed accordingly. In our project, we chose laccase and dioxygenase for BaP degradation. We also planned to test the system with bimolecular fluorescence complementation (BiFC) dimerization. | |
| - | + | To become functional, the PDZ domain and PDZ ligand bind the two protein fragments together, forming a stable dimer.</p> | |
| - | + | <p><h3>2. Proposed Mechanism of Switching “ON” and “OFF” in the System</h3></p> | |
| - | + | <p><strong>Switching “OFF”</strong><br> | |
| - | + | When no external charge is applied, the two effectors are combined, as the repulsion force between the two positively-charged voltage sensor peptide is relatively smaller than the dimer effect. The two voltage sensor peptides are vertically parallel to each other.</p> | |
| - | + | <p align="center"><img src="https://static.igem.org/mediawiki/2013/0/0f/Vs2.png" alt="3" width="472" height="288"></p> | |
| - | + | <p><strong>Switching “ON”</strong><br> | |
| - | <p> | + | When negative charge is applied into the system, the two effectors are separated. As the potential across the membrane is inverted, there is an attractive force between negatively-charged periplasm and positively-charged voltage sensor peptide, and vice versa for cytosol and voltage sensor peptide. Together, these two forces pull the two voltage sensor peptides from vertical position to more horizontal, which causes the separation of the effectors.</p> |
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2013/d/d3/Vs3.png" alt="4" width="482" height="292"><br clear="all"> | ||
| + | </p> | ||
| + | |||
</p2> | </p2> | ||
</div> | </div> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 162: | Line 169: | ||
Address: Rm. 184, Science Centre, CUHK <br/> | Address: Rm. 184, Science Centre, CUHK <br/> | ||
Email: kingchan@cuhk.edu.hk Tel: (852)-39434420 Fax: (852)-26037246 | Email: kingchan@cuhk.edu.hk Tel: (852)-39434420 Fax: (852)-26037246 | ||
| - | + | <div id="backtop"><a href="#">TOP</a></div></div> | |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 03:57, 29 October 2013
 "
"