Team:Hong Kong CUHK/backgroundVS
From 2013.igem.org
(Difference between revisions)
Janeytling (Talk | contribs) |
Janeytling (Talk | contribs) |
||
| (2 intermediate revisions not shown) | |||
| Line 72: | Line 72: | ||
position:relative; | position:relative; | ||
width:1100px; | width:1100px; | ||
| - | height: | + | height:1900px; |
z-index:1; | z-index:1; | ||
background-color: #FFF; | background-color: #FFF; | ||
| Line 79: | Line 79: | ||
position:relative; | position:relative; | ||
width:1025px; | width:1025px; | ||
| - | height: | + | height:1900px; |
z-index:1; | z-index:1; | ||
background-color: #FFF; | background-color: #FFF; | ||
| Line 135: | Line 135: | ||
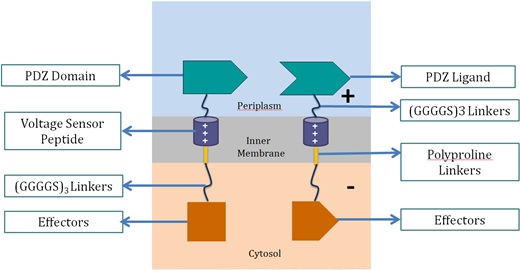
<p>Voltage Switch is a response system triggered by a change of potential across the bacterial inner membrane. Components of this system include the transmembrane voltage sensor peptide originated from potassium ion channels, polyproline linker in the inner membrane, glycine serine linker, PDZ domain and PDZ ligand in the periplasm, and glycine serine linker and effectors in the cytosol.</p> | <p>Voltage Switch is a response system triggered by a change of potential across the bacterial inner membrane. Components of this system include the transmembrane voltage sensor peptide originated from potassium ion channels, polyproline linker in the inner membrane, glycine serine linker, PDZ domain and PDZ ligand in the periplasm, and glycine serine linker and effectors in the cytosol.</p> | ||
<h3>1. Components of Voltage Switch System</h3><p align="center"> | <h3>1. Components of Voltage Switch System</h3><p align="center"> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2013/8/84/Vs1.png" alt="2" width="521" height="270"> <br> | |
</p> | </p> | ||
<p><stong> | <p><stong> | ||
| Line 149: | Line 149: | ||
These effectors can be changed accordingly. In our project, we chose laccase and dioxygenase for BaP degradation. We also planned to test the system with bimolecular fluorescence complementation (BiFC) dimerization. | These effectors can be changed accordingly. In our project, we chose laccase and dioxygenase for BaP degradation. We also planned to test the system with bimolecular fluorescence complementation (BiFC) dimerization. | ||
To become functional, the PDZ domain and PDZ ligand bind the two protein fragments together, forming a stable dimer.</p> | To become functional, the PDZ domain and PDZ ligand bind the two protein fragments together, forming a stable dimer.</p> | ||
| - | <p><h3>2. Proposed Mechanism of Switching “ON” and “OFF” in the System<h3></p> | + | <p><h3>2. Proposed Mechanism of Switching “ON” and “OFF” in the System</h3></p> |
<p><strong>Switching “OFF”</strong><br> | <p><strong>Switching “OFF”</strong><br> | ||
When no external charge is applied, the two effectors are combined, as the repulsion force between the two positively-charged voltage sensor peptide is relatively smaller than the dimer effect. The two voltage sensor peptides are vertically parallel to each other.</p> | When no external charge is applied, the two effectors are combined, as the repulsion force between the two positively-charged voltage sensor peptide is relatively smaller than the dimer effect. The two voltage sensor peptides are vertically parallel to each other.</p> | ||
Latest revision as of 03:57, 29 October 2013
 "
"