Team:Kyoto/projectRNA
From 2013.igem.org
(→Introduction) |
|||
| (152 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
</html> | </html> | ||
<div id="projectRNA"> | <div id="projectRNA"> | ||
| - | + | <div class="texts" style="margin-top: -9px;"> | |
| - | + | =RNA Oscillator= | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <div class="texts" style="margin-top: - | + | |
| - | =RNA | + | |
<div id="introtab"> | <div id="introtab"> | ||
==Introduction== | ==Introduction== | ||
| - | + | ===Motivation=== | |
| - | + | Simulating cell-cell interaction model is too complicated to compute because there is a need to consider not only intracellular condition but also more complex conditions such as positional relationship. | |
| - | + | Then we focused on intracellular condition, and considered what makes this difference between dry work and wet work, and makes modeling and experiment closer. A study of synthetic biology shows an oscillation model which is confirmed in both dry and wet lab.[1] Under this experiment, the effect of cell division which seems to give biggest interference with oscillation cycle can be approximated into zero. Consequently, this circuit is robust enough. From this example, one of the solution to deal with difficulties in reconstructing dry model in wet lab is adoption of robust gene-circuit model in order to ignore the complexity by approximation. However, there are difficulties in choosing factors under the limitation of remaining the robustness of the cycle. We worked on a consisting oscillation circuit which can be closely reproduced by computer simulation. Our goal is generating oscillation cycle in both wet and dry lab. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
<div id="activationtab"> | <div id="activationtab"> | ||
| - | == | + | ===Oscillation=== |
| - | + | We propose following circuit with RNA-RNA interaction as repression mechanism and RNA aptamer-TetR protein interaction as activation mechanism. Fluctuation of factors that effects on a model such as cell division can be approximated into zero because the fluctuation becomes narrower with RNA that is produced or discomposed speedy, we think. We choose Spinach as reporter. | |
| - | + | <html> | |
| - | < | + | <center> |
| + | <iframe id="kyoto_prezi" src="http://prezi.com/embed/eaubz-cct4kd/?bgcolor=ffffff&lock_to_path=0&autoplay=0&autohide_ctrls=0&features=undefined&disabled_features=undefined" width="550" height="400" frameBorder="0"> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/a/a0/Kyoto_RNA_Prezi.png"/> | ||
| + | </iframe> | ||
| + | </center> | ||
| + | </html> | ||
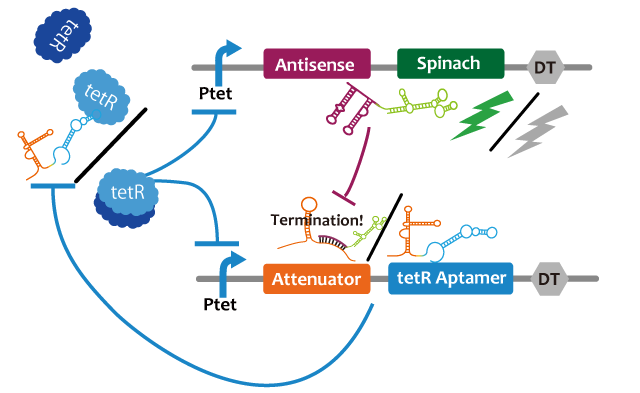
| - | + | This circuit generates oscillation in the following way: Before starting the oscillation, this circuit doesn't generate oscillation due to the repression of attenuator-TetR aptamer by lacI. First, tet promoter(Ptet) is repressed by TetR at the downstream of constitutive promotor. Then, the oscillator is turned on by IPTG. IPTG induces a transcription of TetR aptamer at the downstream of Plac, Spinach, and pT181 antisense at the downstream of Ptet which are transcribed. Because TetR aptamer activates Ptet, positive feedback occurs and more and more TetR aptamer, Spinach, and Antisense are accumulated. Then, this circuit gets fluorescence. After Antisense is accumulated to some extent, TetR aptamer, at the downstream of Attenuator region, is repressed. Then, because new TetR aptamer is not created, the amount of TetR aptamer decreases quickly. Therefore, Ptet is repressed by TetR protein and the amount of Antisense and Spinach falls, too. Then, this circuit loses fluorescence. After the amount of Antisense decreases sufficiently, this circuit recovers first condition. Through this cycle, this circuit acts as an oscillator. | |
| - | + | [[File:Kyoto_RNA_Prezi.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
<div id="reportertab"> | <div id="reportertab"> | ||
| - | == | + | ===Repressor=== |
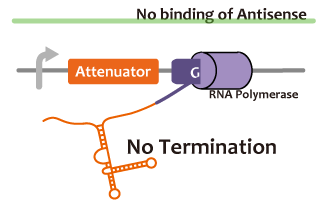
| - | + | We took up non-coding RNA (ncRNA) complementarily binding mRNA as an example of functional RNA which represses transcription. ncRNA in pT181 plasmid (pT181 attenuator) controls the fate of transcriptional elongation in response to an input by complementary antisense RNA. Attenuator region, which lies in 5' untranslated region of a transcript, folds into two different RNA structure. By an interaction with complementary antisense RNA, attenuator region forms Rho-independent terminator and the transcription of the downstream is stopped. Without antisense RNA, attenuator region RNA folds into an alternative structure which allows transcription of the downstream (Novick et al, 1989)[5]. The uniqueness of this mechanism is that it is constructed with only RNA without other small molecules. Synthetic biologists vary functions of RNA only by means of nucleotide substitution etc. (Takahashi et al, 2013)[2]. | |
| - | [ | + | In this paper, many variants of pT181 attenuator/antisense are constructed and the attenuation rate of each variant is different. We chose this mechanism for gene repression. 2013IGKUprojectRNArepressionMECHANISM.png |
| - | + | [[File:2013IGKUprojectRNArepressionMECHANISM.png]] | |
| - | + | [[File:2013IGKUprojectRNArepressionMECHANISM2.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
<div id="repressiontab"> | <div id="repressiontab"> | ||
| - | == | + | ===Activator=== |
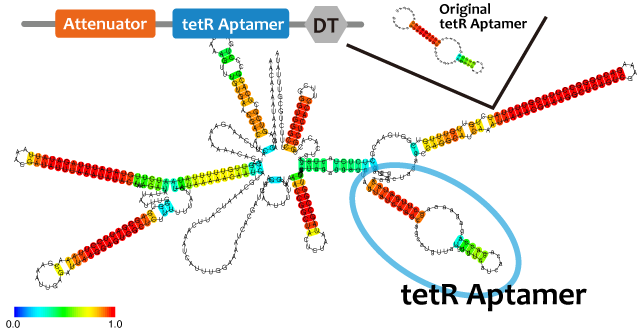
| - | + | We took up TetR aptamer as an example of functional RNA which induces transcription. TetR aptamer induces tetracycline promoter (Ptet) by binding with tetracycline repressor (TetR), which represses Ptet. When TetR aptamer binds to TetR, it induces the conformational change of TetR. As a result, TetR cannot come to bind to tetracycline operator (tetO). We ordered MBL=IDT gene synthesis of pT181 attenuator region DNA, antisense DNA and TetR aptamer with prefix and suffix.We transferred these parts to pSB1C3 and constructed device for antisense and attenuator assay. | |
| - | + | [[File:No-binding-of-tetR-aptamer.png]][[File:Binding-of-tetR-aptamer.png]] | |
</div> | </div> | ||
<div id="fusiontab"> | <div id="fusiontab"> | ||
| + | ===Reporter=== | ||
| + | Spinach is an example of a reporter RNA aptamer,which emits the green fluorescence like GFP when it binds to a fluorophore (DFHBI), which is a derivative fluorophore of GFP. DFHBI doesn't emit fluorescence alone. That is to say, if fluorescence is observed after DFHBI is added into liquid culture, it manifests that Spinach is expressed. If Spinach exists, it combines with DFHBI and DFHBI emits fluorescence. Hence, by using Spinach, it’s possible not only to image RNA directly, but also to reflect the transcription level accurately, which can’t be confirmed via stable protein because RNA is degraded faster than protein. <br> | ||
| + | We strongly suggest Spinach aptamer as a reporter of RNA. | ||
| + | [[File:SPINACHの説明.png]] | ||
| - | ==Fusion== | + | ===Fusion=== |
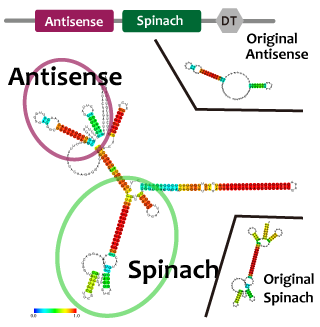
| - | <p> | + | <p>Intending to construct our oscillation circuit, we have to combine two modules into one strand. When we combine two modules, the function of the modules may be inhibited by interactions of secondary structures. In case of RNA, it is relatively easier to predict the morecules' structure. |
| - | + | We estimated the RNA structure to check whether or not unindicatd duplex is formed by open tool. | |
| - | Intending to | + | |
| - | When we combine | + | |
</p> | </p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
<div id="conctab"> | <div id="conctab"> | ||
| - | == | + | ==Experiment== |
| - | + | After we constructed functional RNA generator, we checked the transcription of the RNA parts. To confirm this, we performed RT-PCR.<br> | |
| - | + | samples are following:<br> | |
| + | Negative control<br> | ||
| + | *Non-promoter: Spinach-DT | ||
| + | Experimental group<br> | ||
| + | [[File:唯一のexperiment.png]]<br> | ||
| + | We also checked whether fusion RNA we designed functions or not considering secondary structure with Centroid Fold[6] | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ==Result== | |
| + | ===RT-PCR=== | ||
| + | We performed RT-PCR to confirm transcription of TetR aptamer(left) and Spinach(center).<br> | ||
| + | [[File:ElectrophoresisRT1.png]] | ||
| + | [[File:ElectrophoresisRT2.png]] | ||
| - | + | ===Structure Prediction=== | |
| - | + | [[File:2013IGKUprojectRNAfusionCENTROIDattenuatoraptamer.png]] | |
| + | [[File:antisense_spinach.png]]with Centroid Fold* | ||
<br> | <br> | ||
| - | + | ||
| + | ==Conclusion== | ||
| + | We confirmed the transcription of TetR aptamer, antisense-Spinach, Spinach, and GFP by using RT-PCR method.<br> | ||
| + | We predicted secondary structure of fusion RNA: atenuator-TetRaptamer and antisence-Spinach with centroid fold. It seems to be the expected structure and to function as expected.<br> | ||
| + | We got ready for the construction of the oscilator circuit in wet lab.<br> | ||
| + | <div id="futuretab"> | ||
| + | |||
| + | ==Future work== | ||
| + | To solve simultaneous differential equations meaning oscilation model numerically, we will try to found exact values of some constants. For example, to determine binding constant between TetR and TetR aptamer, we will try to build up assay method and to get quantitative data.<br> | ||
| + | 1. qualitative assay TatR aptamer<br> | ||
| + | To confirm the act of TetR aptamer inducing Ptet ,we are constructing IPTG-inducble TetR aptamer to express GFP. As negative controls, we use RNA with antisense, attenuator, Spinach, no-RNA and attenuator-TetR aptamer. As positive controls, GFP is constitutively expressed.<br> | ||
| + | 3, qualitatively Spinach assay (visual recognition & fluorescence microscopes)<br> | ||
| + | We will check that DFHBI fluorescence on a plate with Spinach.<br> | ||
| + | We will cultivate IPTG-inducible Spinach in a liquid culture under a shading condition, and add DFHBI. Then we check whether this sample fluorescence after centrifugation. We also check Spinach-GFP and antisense-Spinach.<br> | ||
| + | After that, we will substitute the values for oscilation model and try to solve simulate. Moreover we will continue assaying of our parts.<br> | ||
| + | Then, after finishing construction of gene circuits that makes oscilation, we assay the oscilation circuit in wet lab. Our plans for the construction and assay are shown in [https://2013.igem.org/Kyoto:projectRNA/futureview this page]<br> | ||
| + | Finaly, we compare results of wet lab and dry lab and discuss a point in common/difference between the results. | ||
</div> | </div> | ||
<div id="achievetab"> | <div id="achievetab"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
<div id="partslisttab"> | <div id="partslisttab"> | ||
| + | |||
== Parts List == | == Parts List == | ||
| - | |||
| - | |||
<groupparts>iGEM013 Kyoto</groupparts> | <groupparts>iGEM013 Kyoto</groupparts> | ||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
<div id="referencetab"> | <div id="referencetab"> | ||
== Reference == | == Reference == | ||
| - | [ | + | [1][http://www.nature.com/nature/journal/v456/n7221/abs/nature07389.html Jesse Stricker et al.(2008)"A fast, robust and tunable synthetic gene oscillator" Nature 456, 516-519]<br> |
| - | [ | + | [2][http://www.ncbi.nlm.nih.gov/pubmed/23761434 Melissa K. Takahashi and Julius B. Lucks.(2013)"A modular strategy for engineering orthogonal chimeric RNA transcription regulators"Nucleic Acids Research 41(15),7577-88]<br> |
| - | [5 | + | [3][http://www.ncbi.nlm.nih.gov/pubmed/19246008 Anke Hunsicker et al.(2009)"An RNA aptamer that induces transcription"Chem Biol,16(2),173-180]<br> |
| + | [4][http://www.sciencemag.org/content/333/6042/642.abstract Jeremy S. Paige et al.(2011)"RNA Mimics of Green Fluorescent Protein"Science Vol. 333 no. 6042 pp. 642-646]<br> | ||
| + | [5][http://www.ncbi.nlm.nih.gov/pubmed/2478296 Novick RP et al. (1999) "pT181 Plasmid Replication Is Regulated by a Countertranscript-Driven Transcriptional Attenuator"]<br> | ||
| + | [6][http://www.ncrna.org/ Functional RNA Project provided by Computational Biology Research Center (CBRC)]<br> | ||
</div> | </div> | ||
Latest revision as of 12:45, 10 October 2013
count down
Contents |
RNA Oscillator
Introduction
Motivation
Simulating cell-cell interaction model is too complicated to compute because there is a need to consider not only intracellular condition but also more complex conditions such as positional relationship. Then we focused on intracellular condition, and considered what makes this difference between dry work and wet work, and makes modeling and experiment closer. A study of synthetic biology shows an oscillation model which is confirmed in both dry and wet lab.[1] Under this experiment, the effect of cell division which seems to give biggest interference with oscillation cycle can be approximated into zero. Consequently, this circuit is robust enough. From this example, one of the solution to deal with difficulties in reconstructing dry model in wet lab is adoption of robust gene-circuit model in order to ignore the complexity by approximation. However, there are difficulties in choosing factors under the limitation of remaining the robustness of the cycle. We worked on a consisting oscillation circuit which can be closely reproduced by computer simulation. Our goal is generating oscillation cycle in both wet and dry lab.
Oscillation
We propose following circuit with RNA-RNA interaction as repression mechanism and RNA aptamer-TetR protein interaction as activation mechanism. Fluctuation of factors that effects on a model such as cell division can be approximated into zero because the fluctuation becomes narrower with RNA that is produced or discomposed speedy, we think. We choose Spinach as reporter.
This circuit generates oscillation in the following way: Before starting the oscillation, this circuit doesn't generate oscillation due to the repression of attenuator-TetR aptamer by lacI. First, tet promoter(Ptet) is repressed by TetR at the downstream of constitutive promotor. Then, the oscillator is turned on by IPTG. IPTG induces a transcription of TetR aptamer at the downstream of Plac, Spinach, and pT181 antisense at the downstream of Ptet which are transcribed. Because TetR aptamer activates Ptet, positive feedback occurs and more and more TetR aptamer, Spinach, and Antisense are accumulated. Then, this circuit gets fluorescence. After Antisense is accumulated to some extent, TetR aptamer, at the downstream of Attenuator region, is repressed. Then, because new TetR aptamer is not created, the amount of TetR aptamer decreases quickly. Therefore, Ptet is repressed by TetR protein and the amount of Antisense and Spinach falls, too. Then, this circuit loses fluorescence. After the amount of Antisense decreases sufficiently, this circuit recovers first condition. Through this cycle, this circuit acts as an oscillator.
Repressor
We took up non-coding RNA (ncRNA) complementarily binding mRNA as an example of functional RNA which represses transcription. ncRNA in pT181 plasmid (pT181 attenuator) controls the fate of transcriptional elongation in response to an input by complementary antisense RNA. Attenuator region, which lies in 5' untranslated region of a transcript, folds into two different RNA structure. By an interaction with complementary antisense RNA, attenuator region forms Rho-independent terminator and the transcription of the downstream is stopped. Without antisense RNA, attenuator region RNA folds into an alternative structure which allows transcription of the downstream (Novick et al, 1989)[5]. The uniqueness of this mechanism is that it is constructed with only RNA without other small molecules. Synthetic biologists vary functions of RNA only by means of nucleotide substitution etc. (Takahashi et al, 2013)[2].
In this paper, many variants of pT181 attenuator/antisense are constructed and the attenuation rate of each variant is different. We chose this mechanism for gene repression. 2013IGKUprojectRNArepressionMECHANISM.png
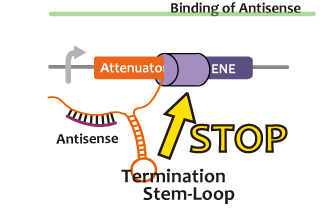
Activator
We took up TetR aptamer as an example of functional RNA which induces transcription. TetR aptamer induces tetracycline promoter (Ptet) by binding with tetracycline repressor (TetR), which represses Ptet. When TetR aptamer binds to TetR, it induces the conformational change of TetR. As a result, TetR cannot come to bind to tetracycline operator (tetO). We ordered MBL=IDT gene synthesis of pT181 attenuator region DNA, antisense DNA and TetR aptamer with prefix and suffix.We transferred these parts to pSB1C3 and constructed device for antisense and attenuator assay.
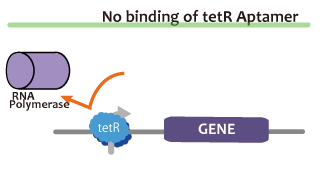
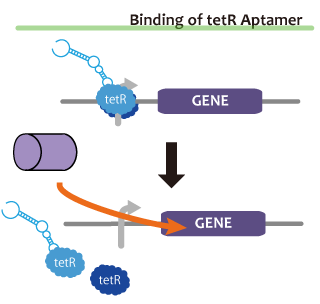
Reporter
Spinach is an example of a reporter RNA aptamer,which emits the green fluorescence like GFP when it binds to a fluorophore (DFHBI), which is a derivative fluorophore of GFP. DFHBI doesn't emit fluorescence alone. That is to say, if fluorescence is observed after DFHBI is added into liquid culture, it manifests that Spinach is expressed. If Spinach exists, it combines with DFHBI and DFHBI emits fluorescence. Hence, by using Spinach, it’s possible not only to image RNA directly, but also to reflect the transcription level accurately, which can’t be confirmed via stable protein because RNA is degraded faster than protein.
We strongly suggest Spinach aptamer as a reporter of RNA.
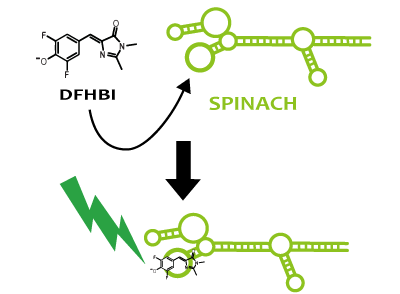
Fusion
Intending to construct our oscillation circuit, we have to combine two modules into one strand. When we combine two modules, the function of the modules may be inhibited by interactions of secondary structures. In case of RNA, it is relatively easier to predict the morecules' structure. We estimated the RNA structure to check whether or not unindicatd duplex is formed by open tool.
Experiment
After we constructed functional RNA generator, we checked the transcription of the RNA parts. To confirm this, we performed RT-PCR.
samples are following:
Negative control
- Non-promoter: Spinach-DT
Experimental group
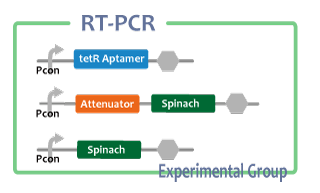
We also checked whether fusion RNA we designed functions or not considering secondary structure with Centroid Fold[6]
Result
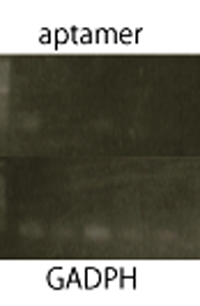
RT-PCR
We performed RT-PCR to confirm transcription of TetR aptamer(left) and Spinach(center).

Structure Prediction
Conclusion
We confirmed the transcription of TetR aptamer, antisense-Spinach, Spinach, and GFP by using RT-PCR method.
We predicted secondary structure of fusion RNA: atenuator-TetRaptamer and antisence-Spinach with centroid fold. It seems to be the expected structure and to function as expected.
We got ready for the construction of the oscilator circuit in wet lab.
Future work
To solve simultaneous differential equations meaning oscilation model numerically, we will try to found exact values of some constants. For example, to determine binding constant between TetR and TetR aptamer, we will try to build up assay method and to get quantitative data.
1. qualitative assay TatR aptamer
To confirm the act of TetR aptamer inducing Ptet ,we are constructing IPTG-inducble TetR aptamer to express GFP. As negative controls, we use RNA with antisense, attenuator, Spinach, no-RNA and attenuator-TetR aptamer. As positive controls, GFP is constitutively expressed.
3, qualitatively Spinach assay (visual recognition & fluorescence microscopes)
We will check that DFHBI fluorescence on a plate with Spinach.
We will cultivate IPTG-inducible Spinach in a liquid culture under a shading condition, and add DFHBI. Then we check whether this sample fluorescence after centrifugation. We also check Spinach-GFP and antisense-Spinach.
After that, we will substitute the values for oscilation model and try to solve simulate. Moreover we will continue assaying of our parts.
Then, after finishing construction of gene circuits that makes oscilation, we assay the oscilation circuit in wet lab. Our plans for the construction and assay are shown in this page
Finaly, we compare results of wet lab and dry lab and discuss a point in common/difference between the results.
Parts List
<groupparts>iGEM013 Kyoto</groupparts>
Reference
[1]Jesse Stricker et al.(2008)"A fast, robust and tunable synthetic gene oscillator" Nature 456, 516-519
[2]Melissa K. Takahashi and Julius B. Lucks.(2013)"A modular strategy for engineering orthogonal chimeric RNA transcription regulators"Nucleic Acids Research 41(15),7577-88
[3]Anke Hunsicker et al.(2009)"An RNA aptamer that induces transcription"Chem Biol,16(2),173-180
[4]Jeremy S. Paige et al.(2011)"RNA Mimics of Green Fluorescent Protein"Science Vol. 333 no. 6042 pp. 642-646
[5]Novick RP et al. (1999) "pT181 Plasmid Replication Is Regulated by a Countertranscript-Driven Transcriptional Attenuator"
[6]Functional RNA Project provided by Computational Biology Research Center (CBRC)
 "
"