Team:Penn State/PromoterProject
From 2013.igem.org
Plant Promoter Project
As plants are still novel organisms for most of synthetic biology, we we are interested in developing methods of control for our projects. Currently the Cauliflower Mosaic Virus 35S promoter is the most widely used plant promoter. In hopes of increasing the availability of plant promoters, our project aims at testing viral promoters due to their relative efficiency, as well as cytoskeletal protein promoters due to their natural abundance. Testing these promoters in parallel with the CaMV 35S will create a plant promoter catalog which can be used for future iGEMers exploration of plant synthetic biology.
Introduction
Synthetic Biology is still a new field with many opportunities for characterization and regulation. The discovery that DNA could be transferred between organisms occurred in 1946, and the first plant was genetically modified in 1983 (1). Since then, most synthetic biology has dealt with bacteria rather than plants. Bacteria are prokaryotic, simple organisms and the procedures for their genetic modification are shorter. Consequently, eukaryotic organisms, especially plants, have an enormous amount of untapped potential (2). Genetic “parts” for plant organisms are limited and often poorly characterized. Under-developed parts prevent many great scientific ideas for plants from ever getting off the ground.

Among the limitations of working with plants Penn State iGEM noticed that only one promoter was commonly available for expression of proteins in plants: CMV 35s promoter. Since in Eukaryotic organisms the promoter is currently the primary source of variable expression, a wider variety of characterized promoters would be very useful. Penn State iGEM aims to modestly characterize several plant promoters for enhanced expression regulation in plant synthetic biology.
REFERENCES:
- http://www.diffen.com/difference/Eukaryotic_Cell_vs_Prokaryotic_Cell
- http://www.nature.com/scitable/blog/bio2.0/modular_transcription_factors_for_eukaryotic
- http://en.wikipedia.org/wiki/Promoter_%28genetics%29
Background
Eukaryotic organisms tend to be larger more complex units than prokaryotes. Some of the differences include the presence of a nucleus, many cytoskeletal elements, and certain genetic devices (1). The diagram below shows the difference from a synthetic biology perspective (2).
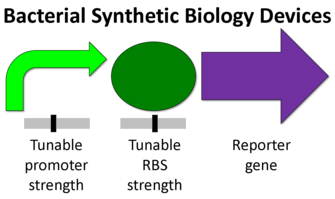
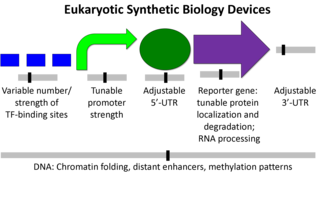
Transcriptions factors, chromosome insertion, the 5- UTR, and the reporter gene itself all can play a role in the protein expression. Although, the promoter region might be the most common and simplest way to modify protein expression in eukaryotes.
Although simple to use, Eukaryotic promoters are fairly complicated. Their function is to call on a RNA polymerase to initiate transcription. The ability of a promoter region to call in an RNA polymerase will determine its strength and the relative protein expression. The are a number of factors involved in this process ,but it varies from promoter to promoter.

REFERENCES:
- http://en.wikipedia.org/wiki/Genetically_modified_food
- http://www2.warwick.ac.uk/fac/sci/lifesci/news/synbio/
Method
...
Results
...
Discussion
...
Further Study
...
Home
Team
Notebook
Promoter Project
Cas9 Project
CesA Project
Butanol Project
Vanillin Project
Parts
Human Practices
Attributions
 "
"