Team:Stanford-Brown/Projects/CRISPR
From 2013.igem.org
Sophia liang (Talk | contribs) (→Additional Applications) |
(→BioBricks) |
||
| Line 38: | Line 38: | ||
== '''BioBricks''' == | == '''BioBricks''' == | ||
| - | '''[http://parts.igem.org/Part:BBa_K1218011 | + | '''[http://parts.igem.org/Part:BBa_K1218011 BBa_K1218011 (pCas9)]''' This part codes for the tracrRNA, Cas9 protein, and minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9. |
| - | '''[http://parts.igem.org/Part:BBa_K1218019 | + | '''[http://parts.igem.org/Part:BBa_K1218019 BBa_K1218019 (CASCADE Complex)]''' This polycistronic sequence contains the CasABCDE genes that form the CASCADE complex. Functions in the innate CRISPR-Cas immune system of Escherichia coli. |
| - | '''[http://parts.igem.org/Part:BBa_K1218014 | + | '''[http://parts.igem.org/Part:BBa_K1218014 BBa_K1218014 (dCas9-ω Activator)]''' This part codes for the tracrRNA, a mutated Cas9 protein, and minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9. However, the Cas9 protein is mutated or "dead" and is fused to a RNAP omega ("w") subunit. Binding of the Cas9-RNAP-subunit fusion activates transcription of the targeted gene. |
== '''Acknowledgements'''== | == '''Acknowledgements'''== | ||
Revision as of 14:46, 27 September 2013
Contents |
Introduction
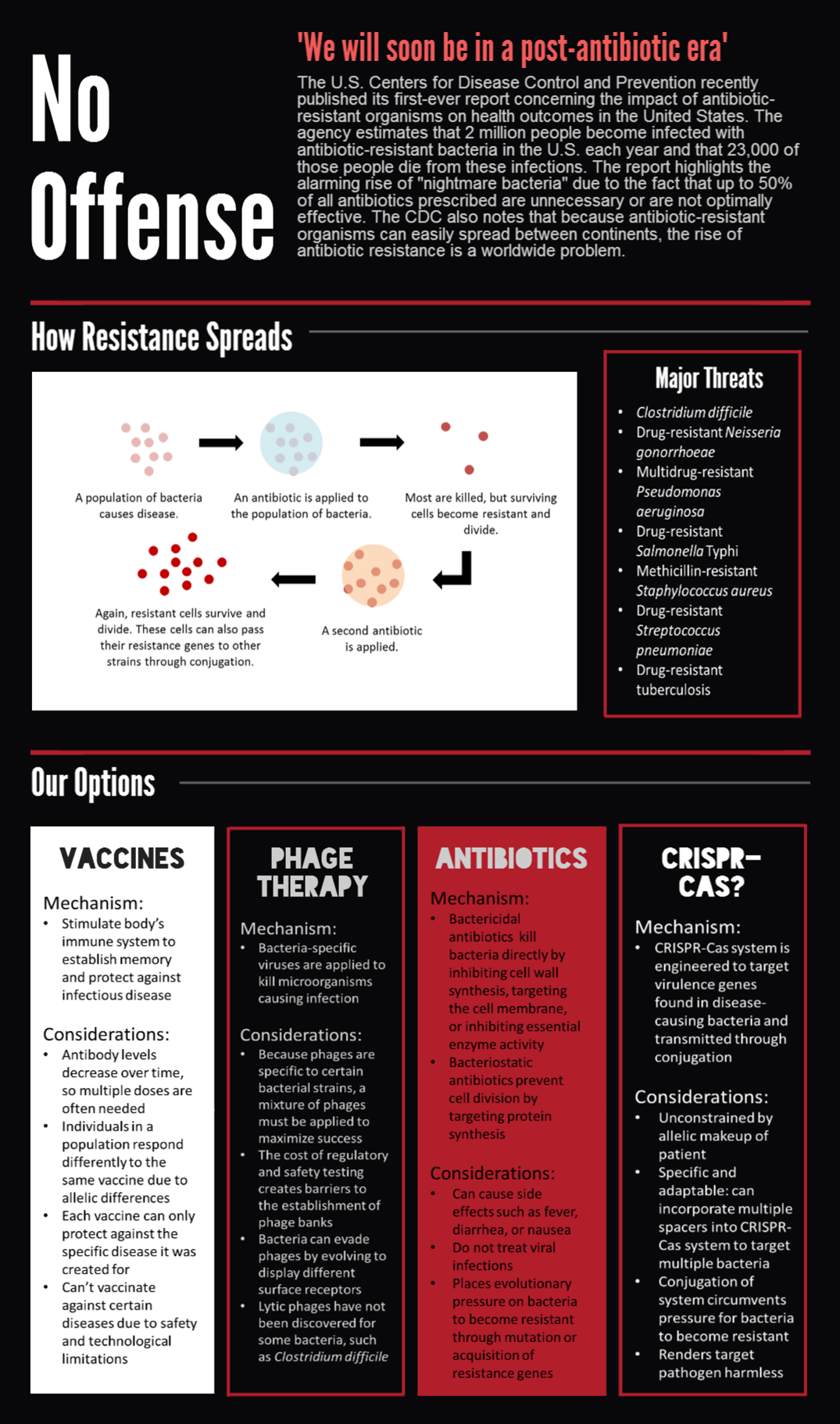
CRISPR-Cas is a bacterial immune system that remembers and targets foreign viral DNA by storing short DNA sequences, called spacers, in arrays called CRISPRs. RNA transcripts of the spacers are then used to sense homologous DNA, which is cleaved by CRISPR-associated (Cas) proteins.
The most commonly studied CRISPR-Cas system is the Cas9 protein from Streptococcus pyogenes, which has drawn much attention for its utility in genome editing. We sought to BioBrick the Cas9 protein and variants for a different purpose — programmable transcriptional regulation. By conjugating a plasmid containing Cas9 and a targeting CRISPR, we aim to regulate specific target genes of an entire population for therapeutic applications.
Protocols
A complete list of our protocols can be found here.
Lab Notebook
Data
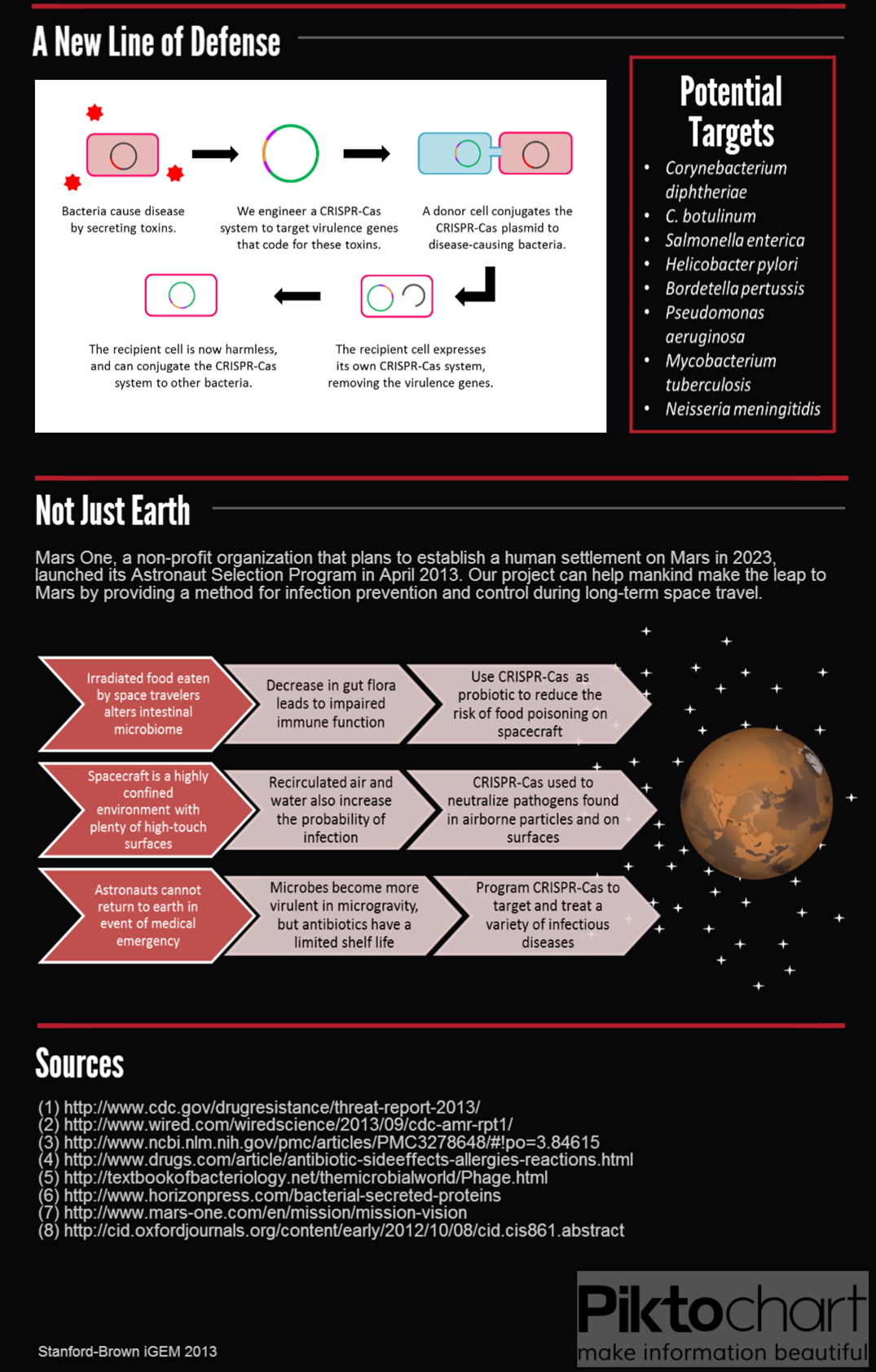
We submitted bricks for Cas9, a minimal CRISPR array, and transcriptional activator variant of Cas9, dCas9-ω. Our system constitutes the first complete, BioBrick compatible CRISPR-Cas system submitted to the registry. Our characterization of the bricks is pending while we construct fluorescent testing constructs to demonstrate the cleaveage activity of Cas9 and regulatory activity of dCas9-ω when introduced via conjugation.
We also demonstrated successful conjugation of the RP4 plasmid. Electrocompetent cells were transformed with RP4, a plasmid coding for proteins needed for conjugation, and grown in liquid culture containing amp. The amp-resistant RP4 cells were mixed with cells containing chlor-resistant RFP, and the resulting strain was plated on a double antibiotic amp+chlor plate.
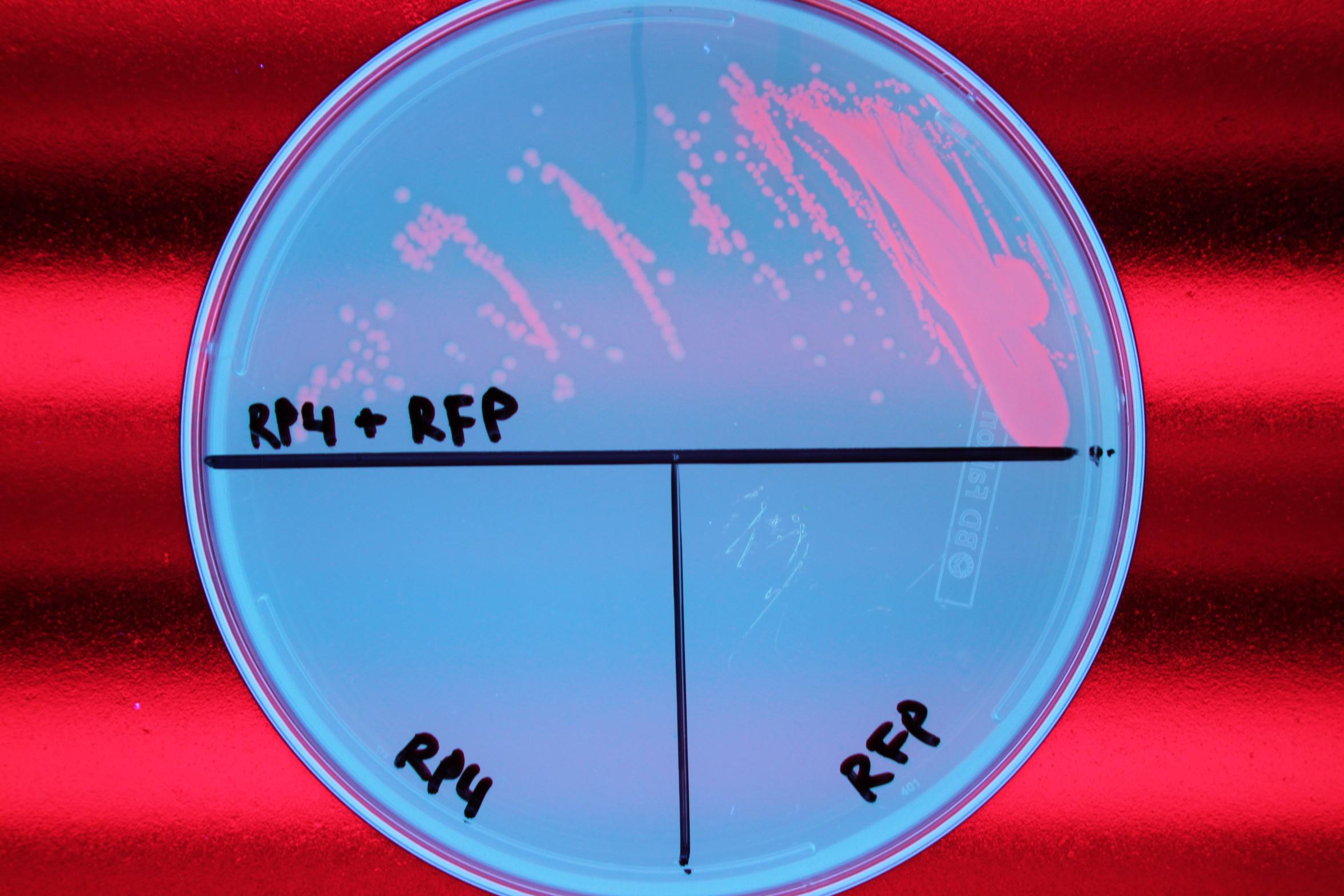
After conjugation was confirmed, we performed a midiprep of the RP4 cells to obtain the RP4 plasmid. Later, this RP4 plasmid will be transformed into our donor cells to facilitate conjugation of the CRISPR-Cas system.
Additional Applications
BioBricks
BBa_K1218011 (pCas9) This part codes for the tracrRNA, Cas9 protein, and minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9.
BBa_K1218019 (CASCADE Complex) This polycistronic sequence contains the CasABCDE genes that form the CASCADE complex. Functions in the innate CRISPR-Cas immune system of Escherichia coli.
BBa_K1218014 (dCas9-ω Activator) This part codes for the tracrRNA, a mutated Cas9 protein, and minimal CRISPR array of a type II CRISPR-Cas system. The CRISPR array includes two CRISPR repeats separated by a spacer with two BsaI sites. Digestion with BsaI allows for insertion of a new spacer, thus changing the sequence targeted by Cas9. However, the Cas9 protein is mutated or "dead" and is fused to a RNAP omega ("w") subunit. Binding of the Cas9-RNAP-subunit fusion activates transcription of the targeted gene.
Acknowledgements
We would like to thank:
- Dr. Lynn Rothschild, Dr. Gary Wessel, and Dr. Joe Shih: primary advisors
- Dr. David Bikard1 and Luciano A. Marraffini: providing Cas9 plasmids and clarifying questions about promoter choice
 "
"