Team:Stanford-Brown/Projects/De-Extinction
From 2013.igem.org
m |
(→Acknowledgements) |
||
| (83 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Stanford-Brown/Templates/Main}} | {{:Team:Stanford-Brown/Templates/Main}} | ||
| - | + | == '''Accomplishments''' == | |
| - | == ''' | + | |
| - | |||
| - | + | ===Ancestral Gene Resurrection=== | |
| + | * Predicted, synthesized, and tested ancestral genes for amino acid synthesis [[File:EvolutionOfLife.png|450px|right|middle]] | ||
| - | + | * Collaborated with Dr. Rich Lenski to test our ancestral gene prediction methods | |
| - | + | ===Submitted 7 BioBricks=== | |
| + | * BBa_K1218003 | ||
| + | * BBa_K1218004 | ||
| + | * BBa_K1218005 | ||
| + | * BBa_K1218006 | ||
| + | * BBa_K1218007 | ||
| + | * BBa_K1218008 | ||
| + | * BBa_K1218009 | ||
| + | __NOTOC__ | ||
| - | + | == '''Introduction''' == | |
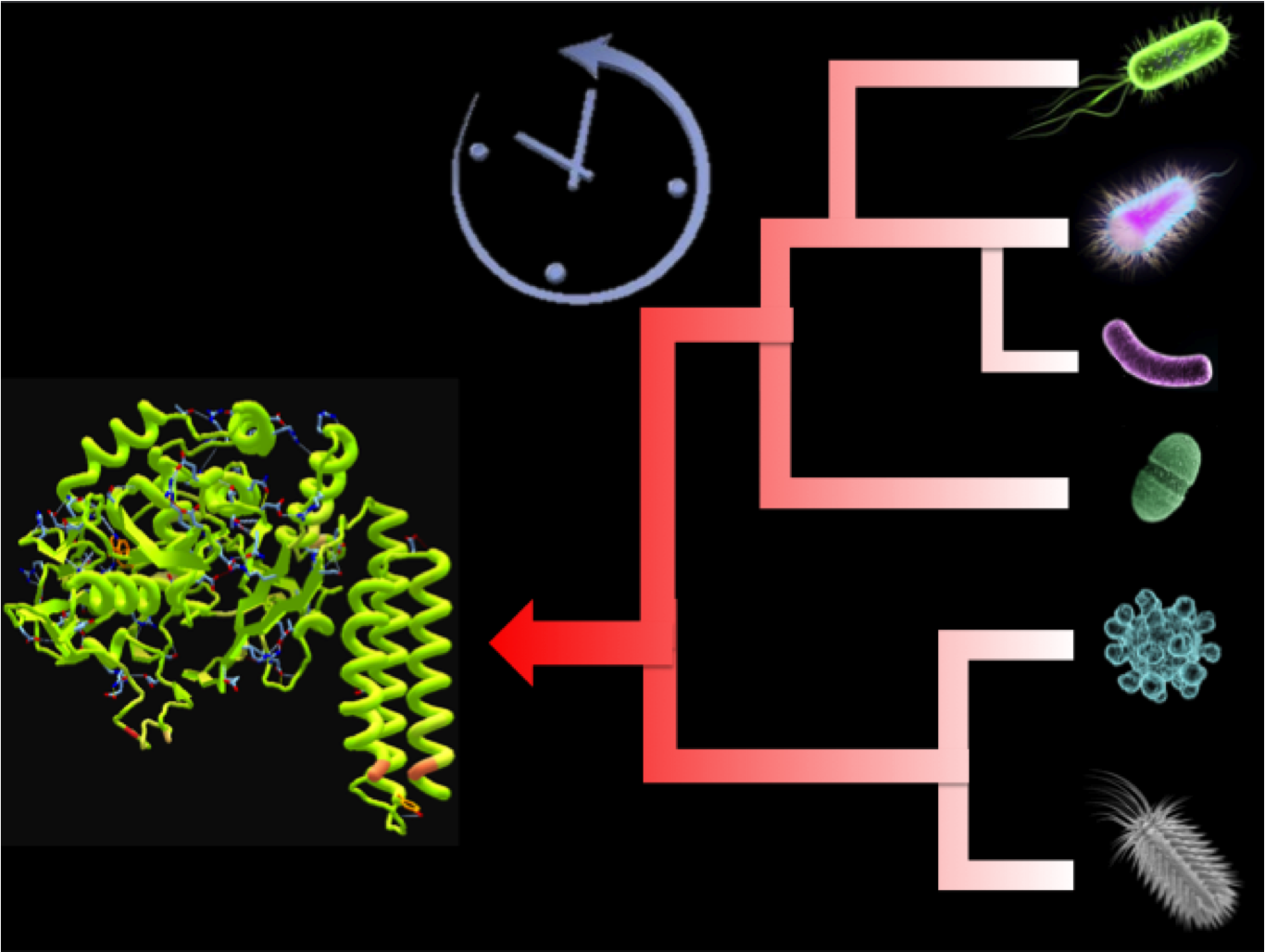
| - | + | [[File:DeX_process.png|300px|thumb|DeExtinction: Turning Back the Phylogenetic Clock]]The Rosetta Stone connected a language well known in modern times to Ancient Egyptian, allowing researchers to better understand Ancient Egypt through their writings. Similarly, the De-Extinction Project seeks to connect our knowledge today to decode messages from the past. We have used phylogenetic analysis by maximum likelihood (PAML) bioinformatic software to predict and reconstruct ancestral genes. This provides a gateway to better understand early life on Earth. | |
| + | ===Amino Acids=== | ||
| - | The De-Extinction | + | Nearly all organisms today uses a standard 20 amino acid alphabet for protein production. However, seven of these did not exist in nature until living organisms began to produce them. The genes CysE and HisC code for proteins used in the production of cysteine and histidine respectively. However, these proteins contain the amino acids that they make, generating a chicken-and-egg question. By reconstructing these ancestral genes and comparing them to the extant genes, we hope to better understand the origin of the amino acid alphabet. |
| + | |||
| + | ===Process Evaluation=== | ||
| + | |||
| + | The De-Extinction project is based on the idea that we can accurately predict ancestral gene sequences. In order to test our methods, we are collaborating with Dr. Rich Lenski at Michigan State University. Dr. Lenski has conducted an evolutionary experiment on E coli for the past 25 years. We are using the most recent sequence data from his experiment to test the accuracy of our ancestral predictions. His data spans 50,000 generations, and has proved very useful in verifying the accuracy of our predictions. | ||
| + | |||
| + | ===Application - CRISPR=== | ||
| + | |||
| + | Latin is a dead language but still has specific use in taxonomy. Similarly, ancestral proteins can be used to expand on modern functionality. Taking this into account, we decided to expand on our team's CRISPR project, which utilizes the prokaryotic immune system for specific recognition and delivery applications (see the "Intercellular" page). The De-Extinction project reconstructed the gene that codes for CasA, the protein subunit responsible for identifying a specific DNA motif. In doing this, we have expanded the number of available CRISPR tools, allowing a variety of DNA motifs to be targeted. We are currently working on biochemical and functional assays to analyze this parts' affinity characteristics and effectiveness. | ||
== '''Protocols''' == | == '''Protocols''' == | ||
| - | + | The '''wet lab protocols''' for the De-Extinction lab procedures can be found [https://docs.google.com/a/brown.edu/document/d/1fMEHTz_VgJtv7hgmuDhb6MxobMkbdJfWQ16Y0s_0GSc/edit#heading=h.37ubqvq53y57 '''here'''] | |
| + | |||
| + | The '''bioinformatic protocols''' for the De-Extinction ancestral protein modeling can be found [https://docs.google.com/a/brown.edu/document/d/11Ff_qpejrQt5ksBnPeJKdrYYbHLferL_43lKN4FPYuk '''here'''] | ||
| + | |||
| + | Our team has compiled a '''complete protocols and lab techniques''' [https://docs.google.com/a/brown.edu/document/d/10hpfdvthGIWRPrrQCnWNIy3HfinFoK0hlV7VMb5nGHI '''here''']. | ||
== '''Lab Notebook''' == | == '''Lab Notebook''' == | ||
| - | The De-Extinction team recorded experiments and progress throughout the research period in the [https://docs.google.com/a/brown.edu/ | + | The De-Extinction team recorded experiments and progress throughout the research period in the [https://docs.google.com/a/brown.edu/file/d/0B8kfpH44xVpraGRkdzlJRHQ1VTQ/edit?usp=drive_web '''De-Extinction Lab Notebook.'''] |
== '''Data''' == | == '''Data''' == | ||
| - | + | ===Molecular Clock=== | |
| - | Using data from Dr. Lenski's lab, we can find the mutation rate for our genes. Using 12 strains and 25 years of data, we found that each of our target genes demonstrate approximately one mutation every 150 years. This is almost exactly the average rate of mutation expected for a one | + | Using data from Dr. Lenski's lab, we can find the mutation rate for our genes. Using 12 strains and 25 years of data, we found that each of our target genes demonstrate approximately one mutation every 150 years. This is almost exactly the average rate of mutation expected for a one kilobase gene in E. coli. |
| + | E. coli have 20 minute generations, meaning around 26,000 generations per year. | ||
| + | [[File:Gyear.jpg]] | ||
| - | + | They typically exhibit 0.001 mutations across the entire genome each generation. | |
| - | + | ||
| - | + | [[File:Myear.jpg]] | |
| - | + | For a gene that makes up 1/4600 fraction of the genome: | |
| + | [[File:Mgene.jpg]] | ||
| - | ' | + | The Lenski lab data shows about 0.0067 mutations per year in this gene, which is close to this calculated value. The nucleotide sequence of our ancestral CasA nucleotide sequence is 54.3% identical to the modern sequence, meaning 815 nucleotides have been mutated. |
| - | Computers, modeling, and bioinformatics are fun for sure, but we were eager to test our genes out as soon as they were | + | |
| + | [[File:Age.jpg]] | ||
| + | |||
| + | This means that our ancestrally predicted gene is at least 120,000 years old! This means that the species we used to predict this gene likely diverged about 120,000 years ago. By using more divergent species in our analysis, we could trace this gene back even farther. | ||
| + | |||
| + | |||
| + | ===Conserved Regions and Protein Analysis=== | ||
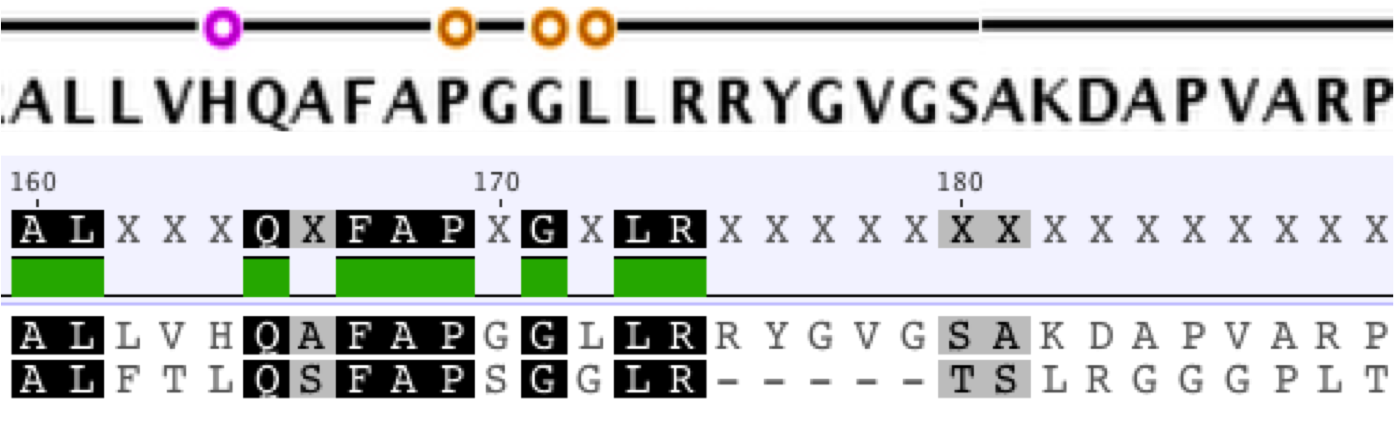
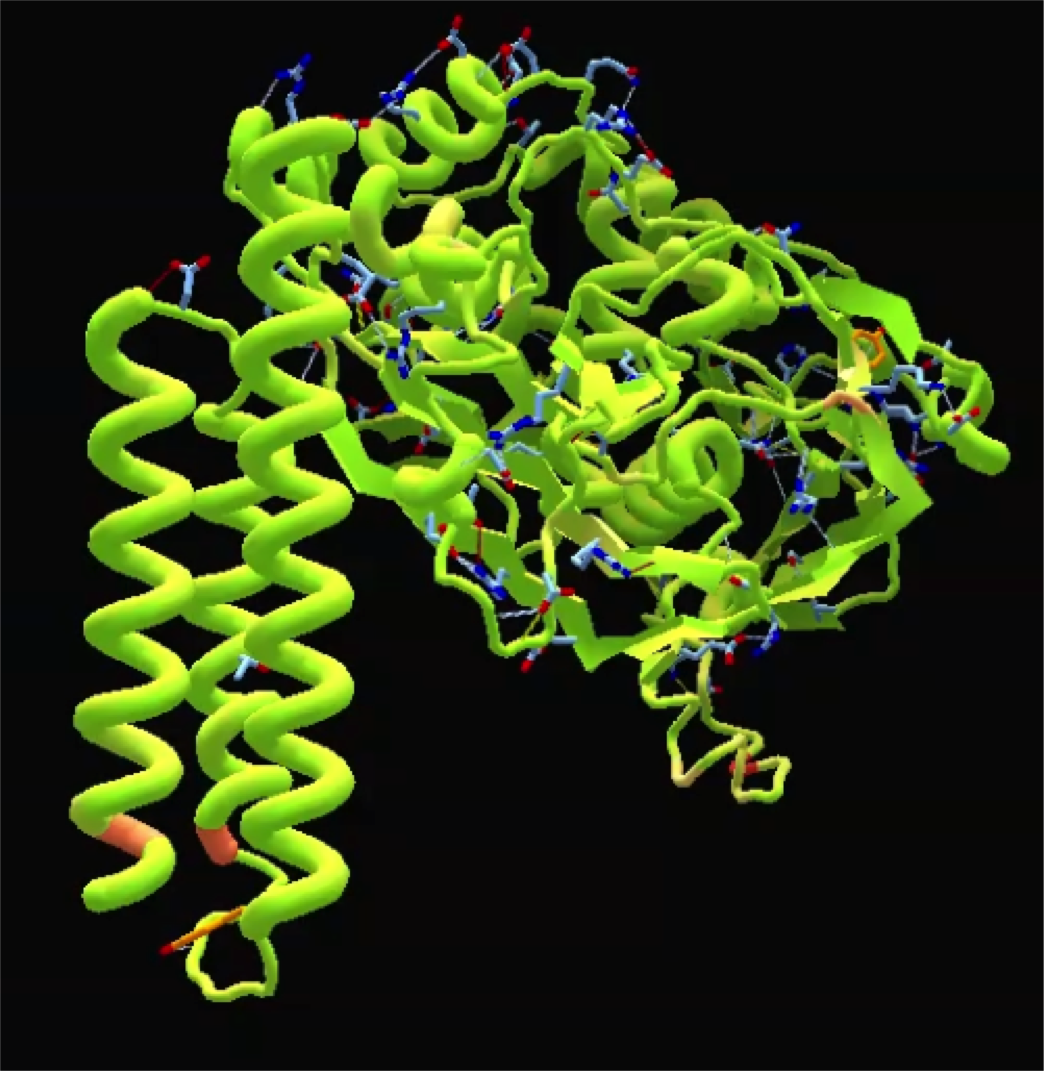
| + | [[File:Conserved_region.png|thumb|left|Conserved Regions Correlate to Binding Sites]][[File:CasA_Foldit_Model.png|thumb|right|A Model of Ancestral CasA]]The extant K12 E. coli HisC and CysE amino acid sequences are 39.1% and 53.3% similar. The amino acid sequences of the extant DH5alpha E. coli casA gene and our ancestral casA gene show 31.1% similarity. The conserved regions of these proteins are not randomly scattered throughout the protein. Instead, the most conserved regions correspond to binding/interaction sites on the protein. As an extension of this observation, we used Foldit software and <i>Thermus thermophilus</i> as a template to make a physical model of our ancestral casA to physically observe the conserved active sites. | ||
| + | |||
| + | We also noticed that, in both HisC and CysE, the number of histidine and cysteine residues, respectively, was smaller in the ancestral proteins. Only one of each residue was conserved between the respective extant and ancestral proteins. This supports a possible solution to the chicken-and-egg problem proposed in the DeExtinction introduction: the these proteins developed first and started making cysteine and histidine before they mutated to incorporate their own products. | ||
| + | |||
| + | After further protein analysis using ExPASy, even more trends with insight about early life were revealed. There are in fact ten amino acids that would not be found in prebiotic contexts: lysine, arginine, tyrosine, phenylalanine, tryptophan, methionine, glutamine, asparagine and aforementioned histidine and cysteine. In most cases, as we would expect, the number of these residues in the ancestral proteins was much smaller than in the extant proteins. Some of these shifts are incredibly dramatic, including the number of asparagine residues in the ancestral CasA fell to 8 from the modern protein that has 32, and glutamine in HisC had 4 residues in the ancestral version from the 21 in the extant protein. This data contributes to build our understanding of the origin of life's 20 amino acid alphabet. | ||
| + | |||
| + | More data that is expected and exciting for our understanding of early life is that each ancestral protein is more hydrophilic than its extant counterpart (grand average of hydropathicity values: extant HisC = -0.035, ancestral HisC= -0.043, extant CysE= 0.002, ancestral CysE= -0.105, extant CasA= -0.193, ancestral CasA= -0.720). Our top theories concerning the origin of life expect life to have originated in water. This supports the idea that the earliest proteins would not have needed a hydrophobic environment to fold properly. Similarly, each ancestral protein has a lower theoretical isoelectric point than its extant counterpart (theoretical isoelectric point: extant HisC = 5.01, ancestral HisC= 4.53, extant CysE= 6.05, ancestral CysE= 5.02, extant CasA= 8.55, ancestral CasA= 6.59). Again, these are results we would expect because it is currently believed that conditions on early Earth were much more acidic than today. Conditions on early Earth were also thought to be much hotter than today as well, and sure enough, ancestral HisC has a higher aliphatic index, which has been shown to be correlated to thermal stability (extant HisC: 101.12, ancestral HisC: 110.22). Furthermore, ancestral HisC was even classified as being more stable than the extant HisC (Instability index (II): extant HisC= 41.71 [unstable], ancestral HisC= 37.83 [stable]). All of this data can be used with modeling programs and experimentation to better understand conditions on early Earth and the rise of early life, but ancestral reconstruction can also be applied to any protein in attempt to make more stable proteins for certain specifications. | ||
| + | |||
| + | Note: For sequence data, refer to the BioBrick pages in the Registry. | ||
| + | |||
| + | ===Wet Lab Testing=== | ||
| + | Computers, modeling, and bioinformatics are fun for sure, but we were eager to test our genes out as soon as they were BioBricks. We used the pUC19 expression vector to express the amino acid production genes we had created/isolated in HisC and CysE knockout cells. We grew these cells on plates of M9 minimal media, so that no amino acids could be picked up from the media. This means that the cells would only survive if they picked up the gene to produce the essential amino acid they could no longer make themselves. We BioBricked the extant HisC gene to use as a positive control for our ancestral HisC tests, and for our ancestral CysE, we used the BioBrick from Trento's 2012 team as a positive control. We used an empty pUC19 plasmid as the negative control for both. Below you can see a zoomed in image of these plates. The colonies on these plates were sequenced to confirm that our ancestral genes successfully rescued their respective cells. It may not be a zombie dinosaur, but we have successfully brought to life something that was extinct long ago! | ||
<gallery> | <gallery> | ||
File:IMG_2870.jpg|''[[Negative HisC control plated on M9 minimal media]]'' (Exhibits few misshaped colonies) | File:IMG_2870.jpg|''[[Negative HisC control plated on M9 minimal media]]'' (Exhibits few misshaped colonies) | ||
File:Negative_CysE_Control.jpg|''[[Negative CysE control plated on M9 minimal media]]'' (Exhibits no colonies) | File:Negative_CysE_Control.jpg|''[[Negative CysE control plated on M9 minimal media]]'' (Exhibits no colonies) | ||
| - | File: | + | File:CysE_Positive_Control.jpg|''[[Positive Control CysE plated on M9 minimal media]]'' (Exhibits many regularly spaced, healthy colonies) |
File:IMG_2857.jpg|''[[Ancestral HisC plated on M9 minimal media]]'' | File:IMG_2857.jpg|''[[Ancestral HisC plated on M9 minimal media]]'' | ||
File:IMG_2864.jpg|''[[Ancestral CysE plated on M9 minimal media]]'' | File:IMG_2864.jpg|''[[Ancestral CysE plated on M9 minimal media]]'' | ||
| Line 52: | Line 92: | ||
== '''Evolutionary Modeling''' == | == '''Evolutionary Modeling''' == | ||
| - | + | ===Background=== | |
| - | + | [[File:Lenski.jpg|thumb|right|One day of the Lesnki Experiment]] | |
Our De-Extinction project was based on the concept of using existing protein and nucleotide information to generate ancestral sequence data. This data would then be synthesized in real life for testing. As a final part of our project, we wanted to evaluate the accuracy of these methods through modeling a known ancestral genome. In order to do this, we collaborated with Dr. Rich Lenski’s lab at Michigan State University. | Our De-Extinction project was based on the concept of using existing protein and nucleotide information to generate ancestral sequence data. This data would then be synthesized in real life for testing. As a final part of our project, we wanted to evaluate the accuracy of these methods through modeling a known ancestral genome. In order to do this, we collaborated with Dr. Rich Lenski’s lab at Michigan State University. | ||
Dr. Lenski has been running a long-term evolutionary experiment on E coli for the past 25 years. In order to test our methods, we gathered recent data from his different strains. This data came from 40,000 generations after the start of the experiment. Because this is not a very long time in evolutionary terms, we focused on mutations across the entire genome, rather than individual significant genes. | Dr. Lenski has been running a long-term evolutionary experiment on E coli for the past 25 years. In order to test our methods, we gathered recent data from his different strains. This data came from 40,000 generations after the start of the experiment. Because this is not a very long time in evolutionary terms, we focused on mutations across the entire genome, rather than individual significant genes. | ||
| - | + | ===Methods=== | |
For our Amino Acid and CRISPR related sequences, we began with amino acid sequence data available from the Pfam (Protein family) Database. We chose to use the seed data, which is representative of the currently known and sequenced population with each specific protein. | For our Amino Acid and CRISPR related sequences, we began with amino acid sequence data available from the Pfam (Protein family) Database. We chose to use the seed data, which is representative of the currently known and sequenced population with each specific protein. | ||
| Line 69: | Line 109: | ||
Because we were examining the entire genome (~4.5 billion bases), using a pairwise aligner was not an option. We instead used Geneious with the Mauve extension, which is designed to accurately align entire genomes. We then used the PhyML extension to generate phylogenetic trees of these strains based on their mutations. | Because we were examining the entire genome (~4.5 billion bases), using a pairwise aligner was not an option. We instead used Geneious with the Mauve extension, which is designed to accurately align entire genomes. We then used the PhyML extension to generate phylogenetic trees of these strains based on their mutations. | ||
| - | We took the aligned nucleotide data and phylogenetic trees as input for PAML with Lazarus. Because this was nucleotide data instead of protein data, we used a nucleotide substitution model | + | We took the aligned nucleotide data and phylogenetic trees as input for PAML with Lazarus. Because this was nucleotide data instead of protein data, we used a nucleotide substitution model. |
| - | + | ===Results=== | |
| - | In the case of the Lenski Lab data, we have the actual ancestral sequence from generation 0. We compared the predicted ancestral sequences to the real ancestral sequence as a test of our methods. The results were over 90% pairwise identical to the real ancestor. | + | [[File:Sample_Alignment.png|300px|thumb|right|An alignment between a predicted sequence and the ancestral sequence]]In the case of the Lenski Lab data, we have the actual ancestral sequence from generation 0. We compared the predicted ancestral sequences to the real ancestral sequence as a test of our methods. The results were over 90% pairwise identical to the real ancestor. As a basic control, we also generated consensus sequences from the same sequences we used to generate our ancestral sequences. The consensus data was 99.9% identical to the ancestral sequences. |
| - | As a basic control, we also generated consensus sequences from the same sequences we used to generate our ancestral sequences. The consensus data was 99.9% identical to the ancestral sequences. | + | |
| + | We have generated data using the Jukes-Cantor 1969 (JC69) model and the Hasegawa, Kishino and Yano 1985 (HKY85) model for comparison. The JC69 model is the most simplistic, assuming equal base frequencies: [[File:Better_base_frequency_identical.png]] and equal mutation rates, leaving the overall mutation rate, μ, as the only parameter. The HKY85 model is much more complex, assuming inequal base frequencies: [[File:Better_unequal_base_frequencies.png]] and unequal mutation rates. It distinguishes between transitions and transversions, making it more complex and more accurate in most instances. | ||
| + | |||
| + | The major differences between these classes can be examined in the rate matrices, which present the likelihood of any base mutating into any other base: | ||
| + | <center> | ||
| + | {| class="wikitable" style="text-align: center" | ||
| + | | [[File:Q_General.png|300px]] | ||
| + | | [[File:Q_JC69.png|250px]] | ||
| + | | [[File:Q_HKY85.png|300px]] | ||
| + | |- | ||
| + | | General Rate Matrix | ||
| + | | Simple JC69 Rate Matrix | ||
| + | | More Complex HKY85 Rate Matrix | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | In our cases, they have yielded similar results. We have seen significant discrepancies, however, between runs where gaps were removed from the data and allowed to form during alignment and runs where gaps were replaced with the most likely nucleotide. In general, removing gaps was superior to replacing gaps. | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 117: | Line 173: | ||
|} | |} | ||
| - | + | ===Discussion=== | |
The preliminary results show that the model is good. 90% identity with the real ancestor is a high mark. But compared to the consensus control, the predicted ancestral sequence is less accurate. | The preliminary results show that the model is good. 90% identity with the real ancestor is a high mark. But compared to the consensus control, the predicted ancestral sequence is less accurate. | ||
| Line 123: | Line 179: | ||
There are multiple reasons why this result may have occurred. It may indicate a problem with the substitution models that we chose to use. For this reason, we are continuing to rerun our data using multiple substitution models for comparison. It may indicate a failure of the Lazarus suite to perform up to expectations when working over short evolutionary periods. The software was designed to predict ancestral sequences of species that have diverged over thousands of years, so using it in a 20 year experiment may be too short of a timeline. It may also indicate a shortcoming in the Lazarus software. | There are multiple reasons why this result may have occurred. It may indicate a problem with the substitution models that we chose to use. For this reason, we are continuing to rerun our data using multiple substitution models for comparison. It may indicate a failure of the Lazarus suite to perform up to expectations when working over short evolutionary periods. The software was designed to predict ancestral sequences of species that have diverged over thousands of years, so using it in a 20 year experiment may be too short of a timeline. It may also indicate a shortcoming in the Lazarus software. | ||
| - | + | ===Future Aims=== | |
| - | + | The potential of ancestral reconstruction is immense. Although our primary use of this technology was creating data to better understand the origin of life and the amino acid alphabet, a scientific endeavor, there are endless engineering projects that can be done. Because early Earth had much hotter and acidic conditions, a reconstruction of almost any protein would most likely be tailored to these conditions. An initial brainstorm of applications that come to mind include prokaryotes that protect buildings and statues from acid rain, plants that are resistant to acid rain, colony screening using heat/acidity, unique bread, beer, or yogurt production procedures, and so much more. As carbon dioxide levels in our atmosphere rise, heat and acidity are becoming global concerns. | |
| - | + | Apart from the thermal and acidic stability of ancestral proteins, aggregating tools with different parameters will diversify the synthetic biology toolbox. For example, an ancestral CRISPR system may not recognize the same type of DNA as modern systems do. With the ancestral CRISPR, we can use two systems with different targets, and activate and silence those systems as we see fit, increasing the control we have over the system. | |
| - | + | With the recognition of its importance, we would like to continue our research into the field of ancestral reconstruction. In the future, we plan to test multiple different substitution models within Lazarus to determine if there is a significant discrepancy in accuracy between them. With access to greater computing power, we could run many more sequences and have more robust results. We also plan to model alternative programs designed for ancestral gene reconstruction. Additionally, we would like to come up with a scenario where we can test these programs over a longer evolutionary period with known ancestral data. This may demonstrate the strengths and weaknesses of these specific algorithms. | |
| + | |||
| + | This only scratches the surface of the evolutionary studies this technology offers. With more time, we could uncover more about the origin of cellular process (e.g. carbon fixation). For the scientific world, we could use cell modeling and ancestral reconstruction in conjunction to understand evolutionary pressures over time. For the engineering world, we could expand the diversity of available tools to include genes and proteins that no longer exist naturally. Eventually, if the reconstruction is accurate enough, who is to say we could not use our own genomes to reconstruct a Neanderthal!? | ||
| + | |||
| + | Be sure to check out our ethics of DeExtinction paper in the [https://2013.igem.org/Team:Stanford-Brown/Projects/HumanPractices '''Human Practices'''] section! | ||
== '''BioBricks''' == | == '''BioBricks''' == | ||
| Line 149: | Line 209: | ||
We would like to thank: | We would like to thank: | ||
| - | - Dr. Lynn Rothschild, Dr. Gary Wessel, Dr. Joe Shih, | + | - Dr. Lynn Rothschild, Dr. Gary Wessel, Dr. Joe Shih, Dr. Kosuke Fujisima, and Diana Gentry |
- Dr. Rich Lenski, the Lenski Lab, and Rohan Maddamsetti, our point of contact | - Dr. Rich Lenski, the Lenski Lab, and Rohan Maddamsetti, our point of contact | ||
| + | |||
| + | == '''References''' == | ||
| + | *Abascal F, Zardoya R, Posada, D. 2005. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics: 21(9):2104-2105. | ||
| + | *Lenski, R. E. (2013). The E. coli long-term experimental evolution project site. http://myxo.css.msu.edu/ecoli | ||
| + | *"New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0." Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. Systematic Biology, 59(3):307-21, 2010. | ||
| + | *The Pfam protein families database: M. Punta, P.C. Coggill, R.Y. Eberhardt, J. Mistry, J. Tate, C. Boursnell, N. Pang, K. Forslund, G. Ceric, J. Clements, A. Heger, L. Holm, E.L.L. Sonnhammer, S.R. Eddy, A. Bateman, R.D. Finn Nucleic Acids Research (2012) Database Issue 40:D290-D301 | ||
| + | *Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in BioSciences 13:555-556. | ||
| + | *Yang, Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586-1591 | ||
Latest revision as of 03:59, 29 October 2013
Accomplishments
Ancestral Gene Resurrection
- Predicted, synthesized, and tested ancestral genes for amino acid synthesis
- Collaborated with Dr. Rich Lenski to test our ancestral gene prediction methods
Submitted 7 BioBricks
- BBa_K1218003
- BBa_K1218004
- BBa_K1218005
- BBa_K1218006
- BBa_K1218007
- BBa_K1218008
- BBa_K1218009
Introduction
The Rosetta Stone connected a language well known in modern times to Ancient Egyptian, allowing researchers to better understand Ancient Egypt through their writings. Similarly, the De-Extinction Project seeks to connect our knowledge today to decode messages from the past. We have used phylogenetic analysis by maximum likelihood (PAML) bioinformatic software to predict and reconstruct ancestral genes. This provides a gateway to better understand early life on Earth.Amino Acids
Nearly all organisms today uses a standard 20 amino acid alphabet for protein production. However, seven of these did not exist in nature until living organisms began to produce them. The genes CysE and HisC code for proteins used in the production of cysteine and histidine respectively. However, these proteins contain the amino acids that they make, generating a chicken-and-egg question. By reconstructing these ancestral genes and comparing them to the extant genes, we hope to better understand the origin of the amino acid alphabet.
Process Evaluation
The De-Extinction project is based on the idea that we can accurately predict ancestral gene sequences. In order to test our methods, we are collaborating with Dr. Rich Lenski at Michigan State University. Dr. Lenski has conducted an evolutionary experiment on E coli for the past 25 years. We are using the most recent sequence data from his experiment to test the accuracy of our ancestral predictions. His data spans 50,000 generations, and has proved very useful in verifying the accuracy of our predictions.
Application - CRISPR
Latin is a dead language but still has specific use in taxonomy. Similarly, ancestral proteins can be used to expand on modern functionality. Taking this into account, we decided to expand on our team's CRISPR project, which utilizes the prokaryotic immune system for specific recognition and delivery applications (see the "Intercellular" page). The De-Extinction project reconstructed the gene that codes for CasA, the protein subunit responsible for identifying a specific DNA motif. In doing this, we have expanded the number of available CRISPR tools, allowing a variety of DNA motifs to be targeted. We are currently working on biochemical and functional assays to analyze this parts' affinity characteristics and effectiveness.
Protocols
The wet lab protocols for the De-Extinction lab procedures can be found here
The bioinformatic protocols for the De-Extinction ancestral protein modeling can be found here
Our team has compiled a complete protocols and lab techniques here.
Lab Notebook
The De-Extinction team recorded experiments and progress throughout the research period in the De-Extinction Lab Notebook.
Data
Molecular Clock
Using data from Dr. Lenski's lab, we can find the mutation rate for our genes. Using 12 strains and 25 years of data, we found that each of our target genes demonstrate approximately one mutation every 150 years. This is almost exactly the average rate of mutation expected for a one kilobase gene in E. coli.
E. coli have 20 minute generations, meaning around 26,000 generations per year.
They typically exhibit 0.001 mutations across the entire genome each generation.
For a gene that makes up 1/4600 fraction of the genome:
The Lenski lab data shows about 0.0067 mutations per year in this gene, which is close to this calculated value. The nucleotide sequence of our ancestral CasA nucleotide sequence is 54.3% identical to the modern sequence, meaning 815 nucleotides have been mutated.
This means that our ancestrally predicted gene is at least 120,000 years old! This means that the species we used to predict this gene likely diverged about 120,000 years ago. By using more divergent species in our analysis, we could trace this gene back even farther.
Conserved Regions and Protein Analysis
The extant K12 E. coli HisC and CysE amino acid sequences are 39.1% and 53.3% similar. The amino acid sequences of the extant DH5alpha E. coli casA gene and our ancestral casA gene show 31.1% similarity. The conserved regions of these proteins are not randomly scattered throughout the protein. Instead, the most conserved regions correspond to binding/interaction sites on the protein. As an extension of this observation, we used Foldit software and Thermus thermophilus as a template to make a physical model of our ancestral casA to physically observe the conserved active sites.We also noticed that, in both HisC and CysE, the number of histidine and cysteine residues, respectively, was smaller in the ancestral proteins. Only one of each residue was conserved between the respective extant and ancestral proteins. This supports a possible solution to the chicken-and-egg problem proposed in the DeExtinction introduction: the these proteins developed first and started making cysteine and histidine before they mutated to incorporate their own products.
After further protein analysis using ExPASy, even more trends with insight about early life were revealed. There are in fact ten amino acids that would not be found in prebiotic contexts: lysine, arginine, tyrosine, phenylalanine, tryptophan, methionine, glutamine, asparagine and aforementioned histidine and cysteine. In most cases, as we would expect, the number of these residues in the ancestral proteins was much smaller than in the extant proteins. Some of these shifts are incredibly dramatic, including the number of asparagine residues in the ancestral CasA fell to 8 from the modern protein that has 32, and glutamine in HisC had 4 residues in the ancestral version from the 21 in the extant protein. This data contributes to build our understanding of the origin of life's 20 amino acid alphabet.
More data that is expected and exciting for our understanding of early life is that each ancestral protein is more hydrophilic than its extant counterpart (grand average of hydropathicity values: extant HisC = -0.035, ancestral HisC= -0.043, extant CysE= 0.002, ancestral CysE= -0.105, extant CasA= -0.193, ancestral CasA= -0.720). Our top theories concerning the origin of life expect life to have originated in water. This supports the idea that the earliest proteins would not have needed a hydrophobic environment to fold properly. Similarly, each ancestral protein has a lower theoretical isoelectric point than its extant counterpart (theoretical isoelectric point: extant HisC = 5.01, ancestral HisC= 4.53, extant CysE= 6.05, ancestral CysE= 5.02, extant CasA= 8.55, ancestral CasA= 6.59). Again, these are results we would expect because it is currently believed that conditions on early Earth were much more acidic than today. Conditions on early Earth were also thought to be much hotter than today as well, and sure enough, ancestral HisC has a higher aliphatic index, which has been shown to be correlated to thermal stability (extant HisC: 101.12, ancestral HisC: 110.22). Furthermore, ancestral HisC was even classified as being more stable than the extant HisC (Instability index (II): extant HisC= 41.71 [unstable], ancestral HisC= 37.83 [stable]). All of this data can be used with modeling programs and experimentation to better understand conditions on early Earth and the rise of early life, but ancestral reconstruction can also be applied to any protein in attempt to make more stable proteins for certain specifications.
Note: For sequence data, refer to the BioBrick pages in the Registry.
Wet Lab Testing
Computers, modeling, and bioinformatics are fun for sure, but we were eager to test our genes out as soon as they were BioBricks. We used the pUC19 expression vector to express the amino acid production genes we had created/isolated in HisC and CysE knockout cells. We grew these cells on plates of M9 minimal media, so that no amino acids could be picked up from the media. This means that the cells would only survive if they picked up the gene to produce the essential amino acid they could no longer make themselves. We BioBricked the extant HisC gene to use as a positive control for our ancestral HisC tests, and for our ancestral CysE, we used the BioBrick from Trento's 2012 team as a positive control. We used an empty pUC19 plasmid as the negative control for both. Below you can see a zoomed in image of these plates. The colonies on these plates were sequenced to confirm that our ancestral genes successfully rescued their respective cells. It may not be a zombie dinosaur, but we have successfully brought to life something that was extinct long ago!
Negative HisC control plated on M9 minimal media (Exhibits few misshaped colonies) |
Negative CysE control plated on M9 minimal media (Exhibits no colonies) |
Positive Control CysE plated on M9 minimal media (Exhibits many regularly spaced, healthy colonies) |
|
Evolutionary Modeling
Background
Our De-Extinction project was based on the concept of using existing protein and nucleotide information to generate ancestral sequence data. This data would then be synthesized in real life for testing. As a final part of our project, we wanted to evaluate the accuracy of these methods through modeling a known ancestral genome. In order to do this, we collaborated with Dr. Rich Lenski’s lab at Michigan State University.
Dr. Lenski has been running a long-term evolutionary experiment on E coli for the past 25 years. In order to test our methods, we gathered recent data from his different strains. This data came from 40,000 generations after the start of the experiment. Because this is not a very long time in evolutionary terms, we focused on mutations across the entire genome, rather than individual significant genes.
Methods
For our Amino Acid and CRISPR related sequences, we began with amino acid sequence data available from the Pfam (Protein family) Database. We chose to use the seed data, which is representative of the currently known and sequenced population with each specific protein.
Using these amino acid sequences, we constructed phylogenetic trees for these specific genes using Geneious with the PhyML extension. We then submitted the sequence and tree data to ProtTest, which we used to determine the most accurate protein substitution model for each protein. In each of our cases the chosen protein model was WAG. Using this data, we ran PAML with the Lazarus suite. We input the amino acid sequence information, phylogenetic tree, and substitution model in order to predict the sequence of the common ancestor of all of the species.
For our real-world evolutionary testing, we altered the process slightly. Dr. Rich Lenski’s lab provided us with nucleotide sequence data for the whole genome in their different strains of E coli.
Because we were examining the entire genome (~4.5 billion bases), using a pairwise aligner was not an option. We instead used Geneious with the Mauve extension, which is designed to accurately align entire genomes. We then used the PhyML extension to generate phylogenetic trees of these strains based on their mutations. We took the aligned nucleotide data and phylogenetic trees as input for PAML with Lazarus. Because this was nucleotide data instead of protein data, we used a nucleotide substitution model.
Results
In the case of the Lenski Lab data, we have the actual ancestral sequence from generation 0. We compared the predicted ancestral sequences to the real ancestral sequence as a test of our methods. The results were over 90% pairwise identical to the real ancestor. As a basic control, we also generated consensus sequences from the same sequences we used to generate our ancestral sequences. The consensus data was 99.9% identical to the ancestral sequences.We have generated data using the Jukes-Cantor 1969 (JC69) model and the Hasegawa, Kishino and Yano 1985 (HKY85) model for comparison. The JC69 model is the most simplistic, assuming equal base frequencies: ![]() and equal mutation rates, leaving the overall mutation rate, μ, as the only parameter. The HKY85 model is much more complex, assuming inequal base frequencies:
and equal mutation rates, leaving the overall mutation rate, μ, as the only parameter. The HKY85 model is much more complex, assuming inequal base frequencies: ![]() and unequal mutation rates. It distinguishes between transitions and transversions, making it more complex and more accurate in most instances.
and unequal mutation rates. It distinguishes between transitions and transversions, making it more complex and more accurate in most instances.
The major differences between these classes can be examined in the rate matrices, which present the likelihood of any base mutating into any other base:
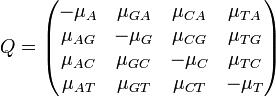
| 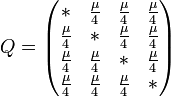
| 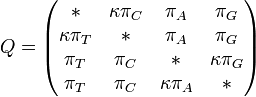
|
| General Rate Matrix | Simple JC69 Rate Matrix | More Complex HKY85 Rate Matrix |
In our cases, they have yielded similar results. We have seen significant discrepancies, however, between runs where gaps were removed from the data and allowed to form during alignment and runs where gaps were replaced with the most likely nucleotide. In general, removing gaps was superior to replacing gaps.
| Genome Segment | Model | Gaps | Percent Similarity to Ancestral Sequence |
|---|---|---|---|
| LCB1 | JC69 | Removed | 26.4% |
| Replaced | 98.6% | ||
| HKY85 | Removed | 98.6% | |
| Replaced | 98.6% | ||
| Consensus | 99.9% | ||
| LCB2 | JC69 | Removed | 99.9% |
| Replaced | 71.4% | ||
| HKY85 | Removed | 99.9% | |
| Replaced | 71.4% | ||
| Consensus | 100% | ||
| LCB3A | JC69 | Removed | 93.7% |
| Replaced | 94.1% | ||
| Consensus | 98.4% | ||
| LCB4 | JC69 | Removed | 99.9% |
| Replaced | 98.8% | ||
| HKY85 | Removed | 99.9% | |
| Replaced | 98.8% | ||
| Consensus | 99.9% |
Discussion
The preliminary results show that the model is good. 90% identity with the real ancestor is a high mark. But compared to the consensus control, the predicted ancestral sequence is less accurate.
There are multiple reasons why this result may have occurred. It may indicate a problem with the substitution models that we chose to use. For this reason, we are continuing to rerun our data using multiple substitution models for comparison. It may indicate a failure of the Lazarus suite to perform up to expectations when working over short evolutionary periods. The software was designed to predict ancestral sequences of species that have diverged over thousands of years, so using it in a 20 year experiment may be too short of a timeline. It may also indicate a shortcoming in the Lazarus software.
Future Aims
The potential of ancestral reconstruction is immense. Although our primary use of this technology was creating data to better understand the origin of life and the amino acid alphabet, a scientific endeavor, there are endless engineering projects that can be done. Because early Earth had much hotter and acidic conditions, a reconstruction of almost any protein would most likely be tailored to these conditions. An initial brainstorm of applications that come to mind include prokaryotes that protect buildings and statues from acid rain, plants that are resistant to acid rain, colony screening using heat/acidity, unique bread, beer, or yogurt production procedures, and so much more. As carbon dioxide levels in our atmosphere rise, heat and acidity are becoming global concerns.
Apart from the thermal and acidic stability of ancestral proteins, aggregating tools with different parameters will diversify the synthetic biology toolbox. For example, an ancestral CRISPR system may not recognize the same type of DNA as modern systems do. With the ancestral CRISPR, we can use two systems with different targets, and activate and silence those systems as we see fit, increasing the control we have over the system.
With the recognition of its importance, we would like to continue our research into the field of ancestral reconstruction. In the future, we plan to test multiple different substitution models within Lazarus to determine if there is a significant discrepancy in accuracy between them. With access to greater computing power, we could run many more sequences and have more robust results. We also plan to model alternative programs designed for ancestral gene reconstruction. Additionally, we would like to come up with a scenario where we can test these programs over a longer evolutionary period with known ancestral data. This may demonstrate the strengths and weaknesses of these specific algorithms.
This only scratches the surface of the evolutionary studies this technology offers. With more time, we could uncover more about the origin of cellular process (e.g. carbon fixation). For the scientific world, we could use cell modeling and ancestral reconstruction in conjunction to understand evolutionary pressures over time. For the engineering world, we could expand the diversity of available tools to include genes and proteins that no longer exist naturally. Eventually, if the reconstruction is accurate enough, who is to say we could not use our own genomes to reconstruct a Neanderthal!?
Be sure to check out our ethics of DeExtinction paper in the Human Practices section!
BioBricks
[http://parts.igem.org/Part:BBa_K1218003 BBa_K1218003 (Modern E. Coli CRISPR CasA)] CasA works in conjunction with the rest of the CASCADE complex as a part of the CRISPR system. Together, the pieces of CASCADE work to recognize, bind to, and degrade foreign DNA.
[http://parts.igem.org/Part:BBa_K1218004 BBa_K1218004 (Ancestral CasA)] CasA works in conjunction with the rest of the CASCADE complex as a part of the CRISPR system. Together, the pieces of CASCADE work to recognize, bind to, and degrade foreign DNA. This part is a predicted sequence that came from a common ancestor of all the species found in the Pfam seed data for CasA (PF09481).
[http://parts.igem.org/Part:BBa_K1218005 BBa_K1218005 (Ancestral CysE)] CysE is responsible for synthesizing cysteine. This part is a fusion of a predicted sequence that came from a common ancestor of all the species found in the Pfam seed data for CysE N-Terminal (PF06426) and the rest of the gene from wild type E coli (K12).
[http://parts.igem.org/Part:BBa_K1218006 BBa_K1218006 (HisC E coli)] HisC is responsible for synthesizing histidine.
[http://parts.igem.org/Part:BBa_K1218007 BBa_K1218007 (Ancestral HisC)] HisC is responsible for synthesizing histidine. This part is a predicted sequence that came from a common ancestor of all the species found in the Pfam seed data for HisC (PF00155).
[http://parts.igem.org/Part:BBa_K1218008 BBa_K1218008 (AroE E coli)] AroE is responsible for synthesizinga shikimate dehydrogenase, which is essential to the production of nucleotides.
[http://parts.igem.org/Part:BBa_K1218009 BBa_K1218009 (CRISPR CasBCDE E. Coli CRISPR)] CasBCDE works in conjunction with CasA in the CASCADE complex as a part of the CRISPR system. Together, the pieces of CASCADE work to recognize, bind to, and degrade foreign DNA.
Acknowledgements
We would like to thank:
- Dr. Lynn Rothschild, Dr. Gary Wessel, Dr. Joe Shih, Dr. Kosuke Fujisima, and Diana Gentry
- Dr. Rich Lenski, the Lenski Lab, and Rohan Maddamsetti, our point of contact
References
- Abascal F, Zardoya R, Posada, D. 2005. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics: 21(9):2104-2105.
- Lenski, R. E. (2013). The E. coli long-term experimental evolution project site. http://myxo.css.msu.edu/ecoli
- "New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0." Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. Systematic Biology, 59(3):307-21, 2010.
- The Pfam protein families database: M. Punta, P.C. Coggill, R.Y. Eberhardt, J. Mistry, J. Tate, C. Boursnell, N. Pang, K. Forslund, G. Ceric, J. Clements, A. Heger, L. Holm, E.L.L. Sonnhammer, S.R. Eddy, A. Bateman, R.D. Finn Nucleic Acids Research (2012) Database Issue 40:D290-D301
- Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in BioSciences 13:555-556.
- Yang, Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586-1591
 "
"