Team:TU-Delft/ProjectOverview
From 2013.igem.org


Methicillin-Resistant Staphylococcus aureus is causing major problems in hospitals and nursing homes. In 2002 it caused over 0.5 million infections in the United States alone, putting patients with open wounds and weakened immune systems at great risks. It is so problematic because it is very difficult to treat; these bacteria have developed resistance against almost all antibiotics available. Several newly discovered strains of MRSA even show antibiotic resistance to vancomycin and teicoplanin, which where one of the last antibiotics useful against MRSA. Although a lot of research is done on alternative treatments, not much success has been booked yet. One of the more promising methods though, seems to be by the use of antimicrobial peptides.
Therefore, our project focusses on designing an E. coli that can detect MRSA in order to locally produce and deliver antimicrobial peptides. Our E. coli will detect S. aureus by its own quorum sensing system, and at that moment inactivated antimicrobial peptide will be produced. With the use of a timer the peptide will be activated by cleaving of an inactivating tag. With the use of the timer, high concentrations of peptide can be delivered very locally in order to efficiently kill MRSA. In order to ensure the safety of the system, the peptide-activating protease is combined with a kill switch that will kill our E. coli (Figure 1).
This project is novel because we aim to fight a life threatening, hard to cure disease by incorporating and combining components of multiple kingdoms into one organism. We let a Gram-negative bacterium sense the communication molecules of gram-positive bacteria and respond to this with an animal innate immune response. The protease activation of the antimicrobial peptide is from fungal origin.
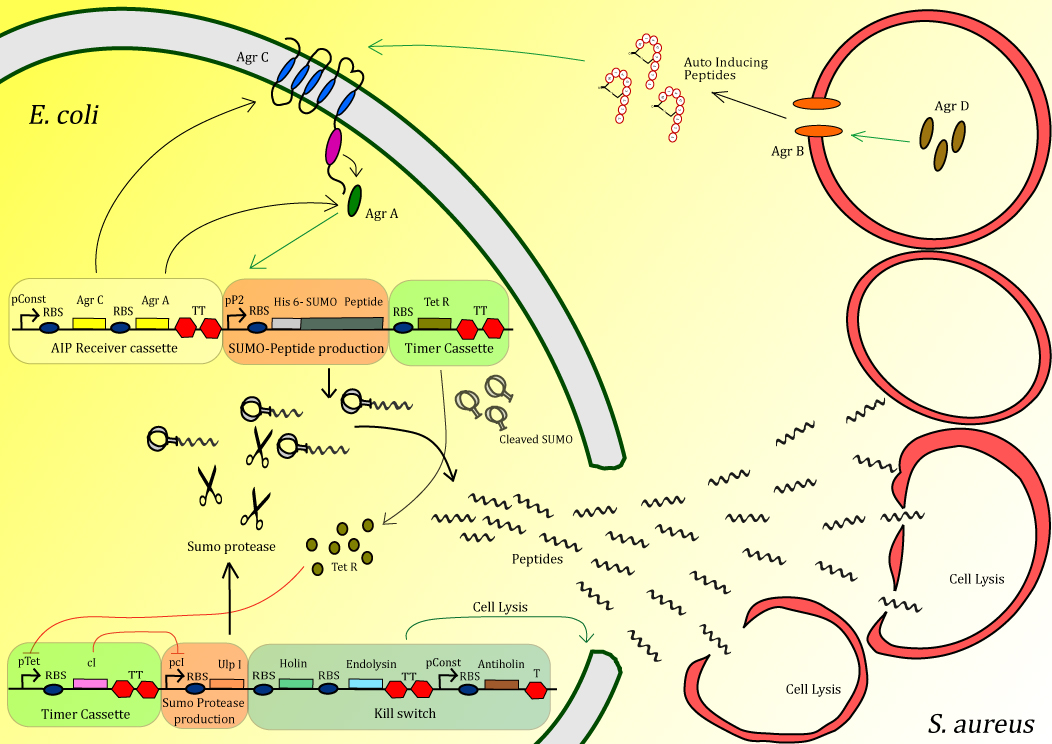
Figure 1: Schematic diagram of the Peptidor system
The project is split up into different modules. Here an overview is given:
Sensing device: The receiving part of the quorum sensing system of S. aureus, AgrA/AgrC operon, is cloned into our Peptidor in order to detect the presence of S. aureus.
Peptide production: After S. aureus has been detected, production of anti-microbial peptide will be started in a manner that allows a strongly localized, high concentration at the exact positition of S. aureus.
Peptide characterization The peptides produced are characterized with the use of both SDS-PAGE and MS/MS.
Timer: The timer was added to the circuit for two reasons. First of all the timer allows Peptidor to build up a high level of peptide before activation through cleavage occurs. When activated too early, the expression host could be severely affected, or the peptides could get trapped in inclusion bodies.
Kill switch : To secure safety for both the patient and the environment, a kill switch is incorporate to ensure lysis of the E. coli expression host.
 "
"
