Team:Tsinghua-E/Project
From 2013.igem.org
| (14 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<style type="text/css"> | <style type="text/css"> | ||
#content{height:1000px;} | #content{height:1000px;} | ||
| - | p{font-size: | + | p{font-size:130%} |
.memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | .memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | ||
.memberx span{z-index:50;position:relative;width:453px;display:block;top:-215px;left:305px;color:#000000} | .memberx span{z-index:50;position:relative;width:453px;display:block;top:-215px;left:305px;color:#000000} | ||
| Line 11: | Line 11: | ||
<ul class="leftdh"> | <ul class="leftdh"> | ||
<li><a href="https://2013.igem.org/Team:Tsinghua-E/Project">Overview</a></li> | <li><a href="https://2013.igem.org/Team:Tsinghua-E/Project">Overview</a></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Tsinghua-E/ | + | <li><a href="https://2013.igem.org/Team:Tsinghua-E/Result">Results</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Tsinghua-E/Film"> | + | <li><a href="https://2013.igem.org/Team:Tsinghua-E/Film">Film</a></li><br/> |
| - | <li><a href=" | + | <li><a href="/Team:Tsinghua-E/Safety">Safety</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 24: | Line 24: | ||
<em>in vivo</em> evolution engineering towards tryptophan-overproduction superbug</h3> | <em>in vivo</em> evolution engineering towards tryptophan-overproduction superbug</h3> | ||
<p> | <p> | ||
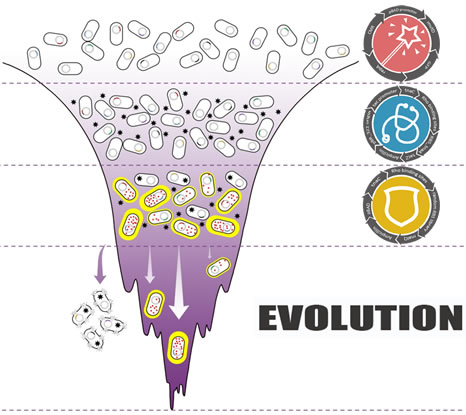
| - | Darwinian evolution shows great power in creating | + | Darwinian evolution shows great power in creating incredible biological function in amazing speed.Inspired by this, our team aimed at creating novel fast and irrational microbial cell factory by simulating natural Darwinian evolution process. With tryptophan as target product, a novel tryptophan biosensor utilizing translating ribosome mechanism was firstly developed as the foundation for tryptophan productivity and selection pressure switch module. We further constructed this tryptophan overproduction selection gene circuit coupling with in vivo mutation machine (mutator gene of mutD). By fine-tuning the selection conditions, our selection circuit showed good tryptophan dependent growth property, which provides the foundation for further evolution.As a preliminary result of this project, we successfully evolved an ancestor with zero productivity to a high-tryptophan producer only after several rounds of evolution. |
</p> | </p> | ||
<h4 align="center"><img src="/wiki/images/5/5f/ThuEproject.jpg" ></h4> | <h4 align="center"><img src="/wiki/images/5/5f/ThuEproject.jpg" ></h4> | ||
Latest revision as of 09:53, 10 October 2013



Overview
Darwinian evolution for microbial cell factory:
in vivo evolution engineering towards tryptophan-overproduction superbug
Darwinian evolution shows great power in creating incredible biological function in amazing speed.Inspired by this, our team aimed at creating novel fast and irrational microbial cell factory by simulating natural Darwinian evolution process. With tryptophan as target product, a novel tryptophan biosensor utilizing translating ribosome mechanism was firstly developed as the foundation for tryptophan productivity and selection pressure switch module. We further constructed this tryptophan overproduction selection gene circuit coupling with in vivo mutation machine (mutator gene of mutD). By fine-tuning the selection conditions, our selection circuit showed good tryptophan dependent growth property, which provides the foundation for further evolution.As a preliminary result of this project, we successfully evolved an ancestor with zero productivity to a high-tryptophan producer only after several rounds of evolution.
 "
"