Team:UC-Santa Cruz/Project/Polarization
From 2013.igem.org
| (9 intermediate revisions not shown) | |||
| Line 12: | Line 12: | ||
</p> | </p> | ||
<p> | <p> | ||
| - | [[File:Seq1.png]] | + | [[File:Seq1.png|600px|center]] |
<br\> | <br\> | ||
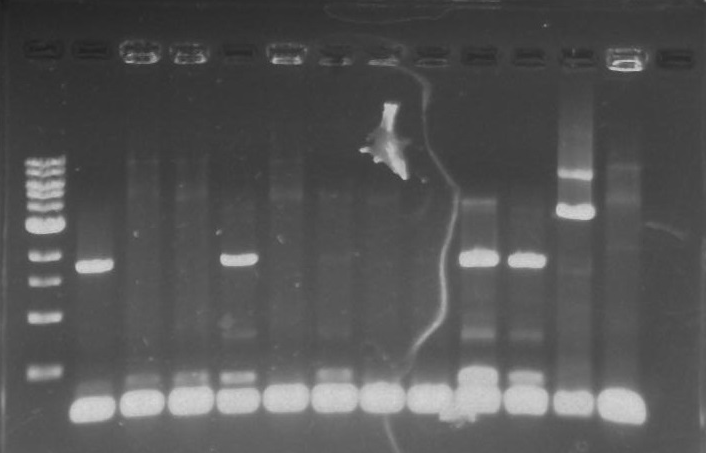
Figure 1. TipF colony PCR Agarose gel. From left to right ladder, TipF1-TipF10, NegControl, PosControl. | Figure 1. TipF colony PCR Agarose gel. From left to right ladder, TipF1-TipF10, NegControl, PosControl. | ||
| Line 21: | Line 21: | ||
</p> | </p> | ||
<p> | <p> | ||
| - | [[File:Dskjfsd.png]] | + | [[File:Dskjfsd.png|600px|center]] |
<br\> | <br\> | ||
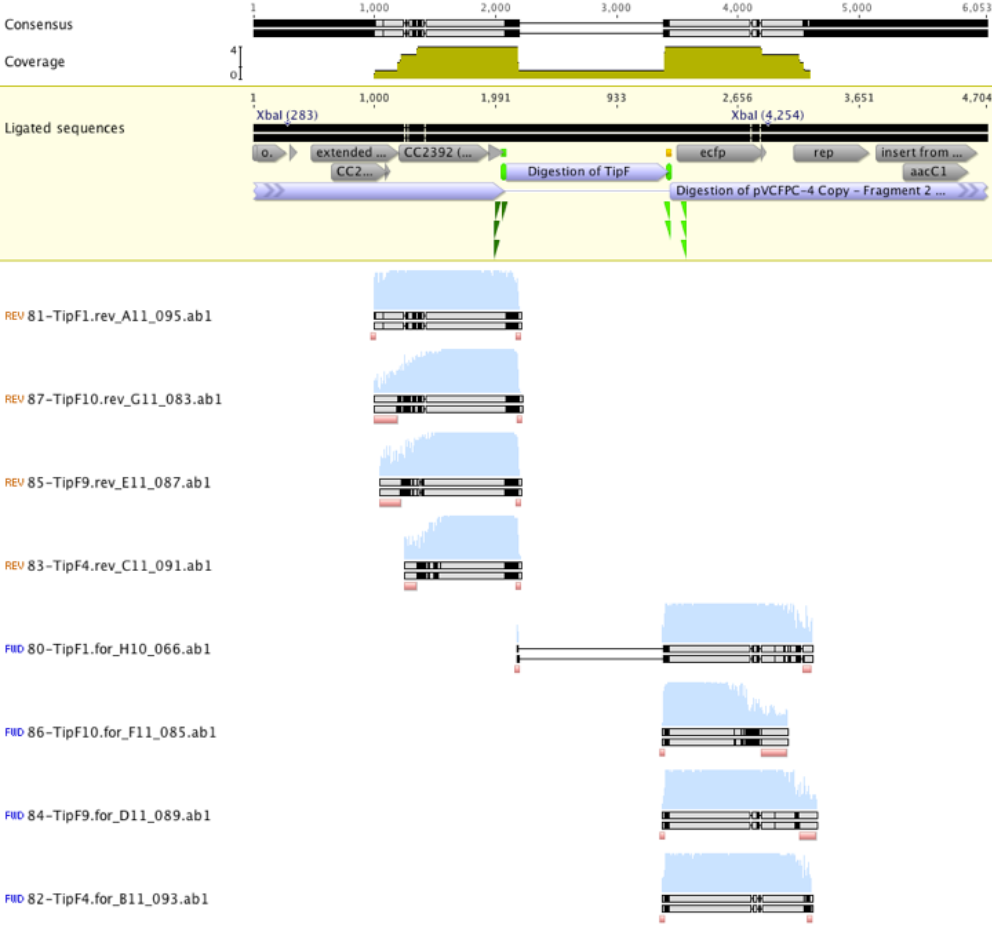
Figure 2. TipF sequencing results aligned to the expected ligation product. Reads suggest sequencing of empty vector. | Figure 2. TipF sequencing results aligned to the expected ligation product. Reads suggest sequencing of empty vector. | ||
| Line 31: | Line 31: | ||
</p> | </p> | ||
<p> | <p> | ||
| + | [[File:Legend.png|600px|center]] | ||
| + | <br\> | ||
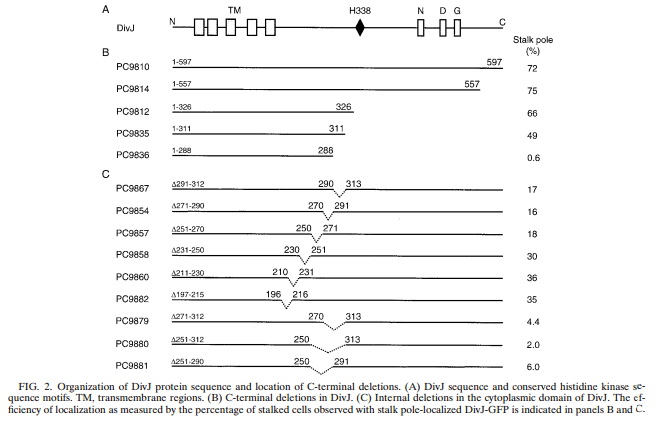
Figure 3. Copied from Sciochetti et al., 2002. | Figure 3. Copied from Sciochetti et al., 2002. | ||
</p> | </p> | ||
| Line 39: | Line 41: | ||
</p> | </p> | ||
<p> | <p> | ||
| + | [[File:Seq2.png|600px|center]] | ||
| + | <br\> | ||
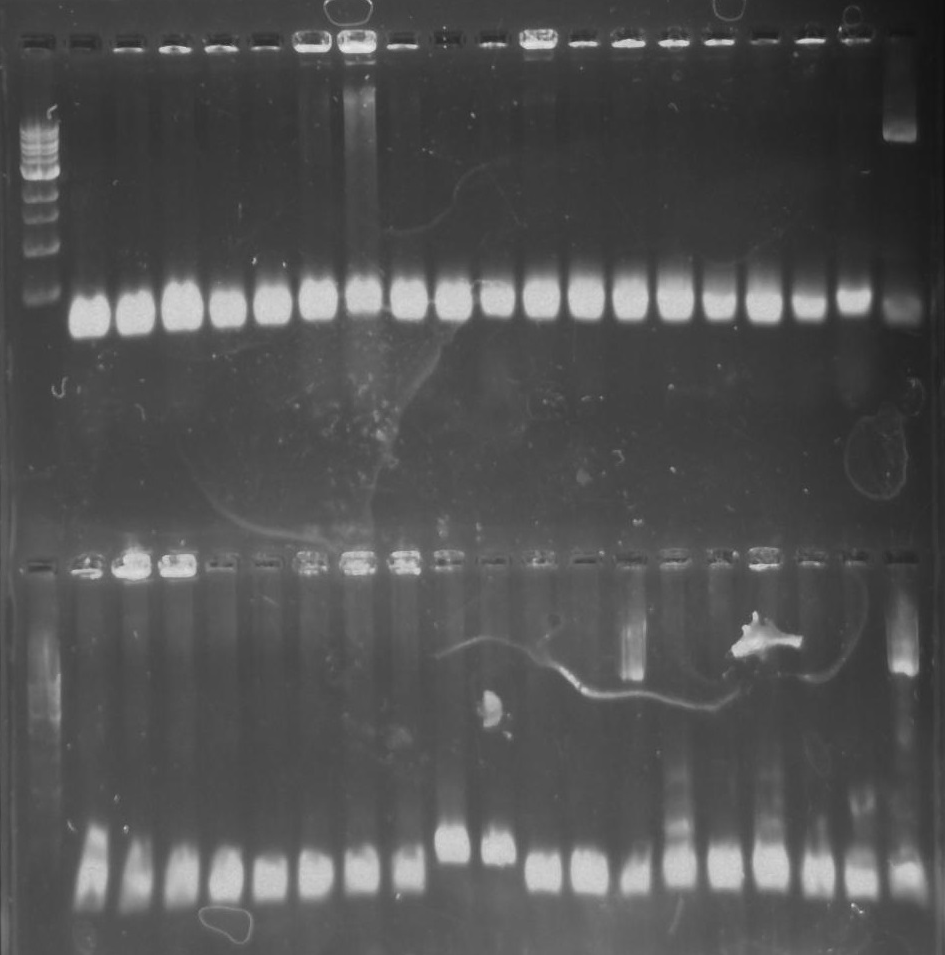
Figure 4. Gel 27: From left to right. Top: Ladder, T1, T2, T3, T4, T5, T6, T7, Y8, T9, T10, CD1, CD2, CD3, CD4, CD5, CD6, CD7, NC, PC. Botton: Ladder, CD8, | Figure 4. Gel 27: From left to right. Top: Ladder, T1, T2, T3, T4, T5, T6, T7, Y8, T9, T10, CD1, CD2, CD3, CD4, CD5, CD6, CD7, NC, PC. Botton: Ladder, CD8, | ||
CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, CA9, CA10, NC, PC, YD1, YD2, YD3, YD4, NY, PY. T = TipF, CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in | CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, CA9, CA10, NC, PC, YD1, YD2, YD3, YD4, NY, PY. T = TipF, CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in | ||
| Line 50: | Line 54: | ||
</p> | </p> | ||
<p> | <p> | ||
| + | [[File:511.png|600px|center]] | ||
| + | <br\> | ||
Figure 5. | Figure 5. | ||
</p> | </p> | ||
<p> | <p> | ||
| + | [[File:DivJMDeletion.png|600px|center]] | ||
| + | <br\> | ||
Figure 6. | Figure 6. | ||
</p> | </p> | ||
| - | + | Citations: | |
| - | + | <br\> | |
| - | + | Dutzler, R. (2004, April 30). Structural basis for ion conduction and gating in ClC chloride channels. FEBS letters. doi:10.1016/S0014-5793(04)00210-8 | |
| - | + | Entcheva-dimitrov, P., Spormann, A. M., Entcheva-dimitrov, P., & Spormann, A. M. (2004). Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter crescentus Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter crescentus, 186(24). doi:10.1128/JB.186.24.8254 | |
| + | <br\> | ||
| + | <br\> | ||
| + | Fields, A. T., Navarrete, C. S., Zare, A. Z., Huang, Z., Mostafavi, M., Lewis, J. C., … Chen, J. C. (2012). The conserved polarity factor podJ1 impacts multiple cell envelope-associated functions in Sinorhizobium meliloti. Molecular microbiology, 84(5), 892–920. doi:10.1111/j.1365-2958.2012.08064.x | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Gitai, Z., Dye, N., & Shapiro, L. (2004). An actin-like gene can determine cell polarity in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 101(23), 8643–8. doi:10.1073/pnas.0402638101 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Hughes, H. V., Huitema, E., Pritchard, S., Keiler, K. C., Brun, Y. V, & Viollier, P. H. (2010). Protein localization and dynamics within a bacterial organelle. Proceedings of the National Academy of Sciences of the United States of America, 107(12), 5599–604. doi:10.1073/pnas.0909119107 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Kwon, S.-K., Kim, B. K., Song, J. Y., Kwak, M.-J., Lee, C. H., Yoon, J.-H., … Kim, J. F. (2013). Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome biology and evolution, 5(1), 187–99. doi:10.1093/gbe/evs134 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Lanyi, J. K., Duschl, A., Hatfield, G. W., May, K., & Wesley, G. (2013). The Primary Structure of a Halorhodopsin Natronobacterium pharaonis from. | ||
| + | Marks, M. E., Castro-Rojas, C. M., Teiling, C., Du, L., Kapatral, V., Walunas, T. L., & Crosson, S. (2010). The genetic basis of laboratory adaptation in Caulobacter crescentus. Journal of bacteriology, 192(14), 3678–88. doi:10.1128/JB.00255-10 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | McCusker, E. C., Bagnéris, C., Naylor, C. E., Cole, A. R., D’Avanzo, N., Nichols, C. G., & Wallace, B. a. (2012). Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nature communications, 3, 1102. doi:10.1038/ncomms2077 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Ng, W. V, Kennedy, S. P., Mahairas, G. G., Berquist, B., Pan, M., Shukla, H. D., … DasSarma, S. (2000, October 24). Genome sequence of Halobacterium species NRC-1. Proceedings of the National Academy of Sciences of the United States of America. doi:10.1073/pnas.190337797 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Obuchowski, P. L., & Jacobs-Wagner, C. (2008). PflI, a protein involved in flagellar positioning in Caulobacter crescentus. Journal of bacteriology, 190(5), 1718–29. doi:10.1128/JB.01706-07 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Ohta, N., Lane, T., Ninfa, E. G., Sommer, J. M., & Newton, a. (1992). A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proceedings of the National Academy of Sciences of the United States of America, 89(21), 10297–301. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=50325&tool=pmcentrez&rendertype=abstract | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Rüdiger, M., & Oesterhelt, D. (1997). Specific arginine and threonine residues control anion binding and transport in the light-driven chloride pump halorhodopsin. The EMBO journal, 16(13), 3813–21. doi:10.1093/emboj/16.13.3813 | ||
| + | Thanbichler, M., Iniesta, A. a, & Shapiro, L. (2007). A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic acids research, 35(20), e137. doi:10.1093/nar/gkm818 | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Sciochetti, Stephen A., et al. "Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus." Journal of bacteriology 184.21 (2002): 6037-6049. | ||
| + | <br\> | ||
| + | <br\> | ||
| + | Zhang, F., Vierock, J., Yizhar, O., Fenno, L. E., Tsunoda, S., Kianianmomeni, A., … Deisseroth, K. (2011). The microbial opsin family of optogenetic tools. Cell, 147(7), 1446–57. doi:10.1016/j.cell.2011.12.004 | ||
Latest revision as of 04:00, 28 September 2013
Polar Tag Results
Isolation of the polar protein tagging genes DivJ, StpX, PflI, and TipF out of a Caulobacter (CB15N) template proved to be more of a challenge than expected. The first round of colony PCR reactions showed several bands at the expected length for the TipF plates. Lanes 1, 4, 9, and 10 showed bright bands at ~1.9 kb (see Figure 1 below)
Figure 1. TipF colony PCR Agarose gel. From left to right ladder, TipF1-TipF10, NegControl, PosControl.
Unfortunately the sequencing results suggested that all reads sequenced were actually empty vector that had been transformed into E.coli (see figure 2 below). A subsequent sequencing of prospective TipF colonies gave similar results.
Figure 2. TipF sequencing results aligned to the expected ligation product. Reads suggest sequencing of empty vector.
After several more similar attempted transformations we decided to build a possible minimal DivJ (DivJM) out of 60 mers (designed using DNA works). We found evidence that suggests a small fragment of DivJ is actually responsible for polar localization of fusion proteins (Sciochetti, 2002)(see figure 3 below).
Figure 3. Copied from Sciochetti et al., 2002.
A ligation reaction and transformation of the recombinant plasmid (with DivJM) into E.coli chemi competent top 10 cells was performed. A colony PCR reaction was performed on the prospective DivJM colonies. Lanes with CA8 and CA9 displayed bright bands at the expected length of ~236 bp (see figure 4 below).
Figure 4. Gel 27: From left to right. Top: Ladder, T1, T2, T3, T4, T5, T6, T7, Y8, T9, T10, CD1, CD2, CD3, CD4, CD5, CD6, CD7, NC, PC. Botton: Ladder, CD8, CA1, CA2, CA3, CA4, CA5, CA6, CA7, CA8, CA9, CA10, NC, PC, YD1, YD2, YD3, YD4, NY, PY. T = TipF, CD = 326 DivJ in pVCPFPC-4. CA = Minimal DivJ in pVCPFPC-4. YD = 326 DivJ in pXYFPC-2. NC = Negative control in pVCPFPC-4, PC = Positive control in pVCPFPC-4, NY = negative control in pXYFPC-2, PY = positive control in pXYFPC-2.
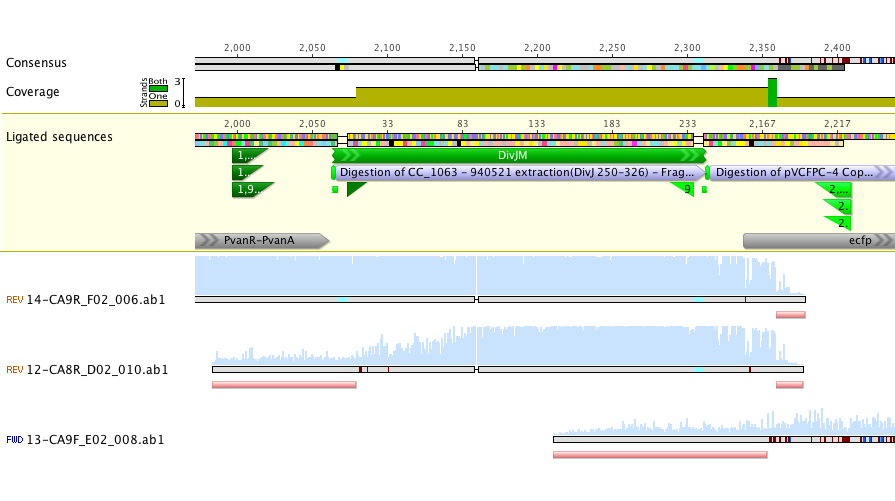
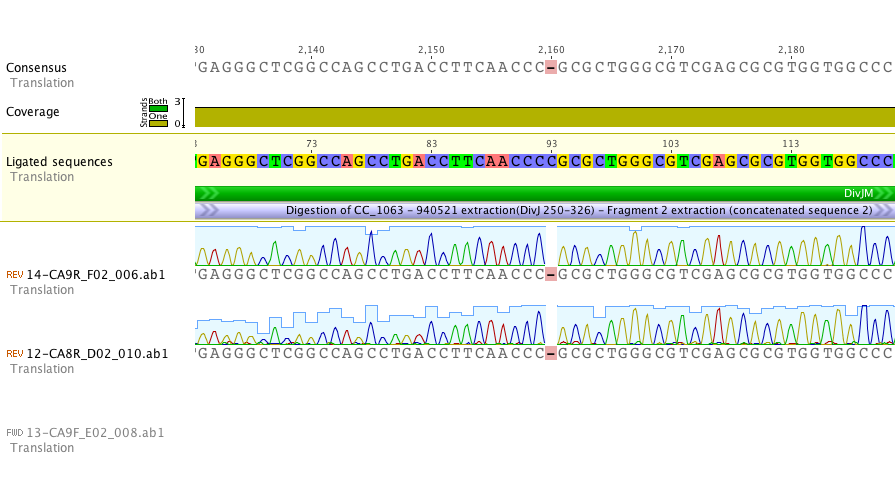
Sequencing of lanes CA8 and CA9 produced reads that suggest a single base deletion was in the DivJM gene (see figure 5 and 6 below). There are several possibilities that would explain the base deletion. An error early on in the synthesis PCR reaction may be to blame for the ambiguous base or there may have been an error in the oligo’s that we had made. We are currently redoing the synthesis and amplification of the DivJM oligo’s.
Figure 5.
Figure 6.
Citations:
Dutzler, R. (2004, April 30). Structural basis for ion conduction and gating in ClC chloride channels. FEBS letters. doi:10.1016/S0014-5793(04)00210-8
Entcheva-dimitrov, P., Spormann, A. M., Entcheva-dimitrov, P., & Spormann, A. M. (2004). Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter crescentus Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter crescentus, 186(24). doi:10.1128/JB.186.24.8254
Fields, A. T., Navarrete, C. S., Zare, A. Z., Huang, Z., Mostafavi, M., Lewis, J. C., … Chen, J. C. (2012). The conserved polarity factor podJ1 impacts multiple cell envelope-associated functions in Sinorhizobium meliloti. Molecular microbiology, 84(5), 892–920. doi:10.1111/j.1365-2958.2012.08064.x
Gitai, Z., Dye, N., & Shapiro, L. (2004). An actin-like gene can determine cell polarity in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 101(23), 8643–8. doi:10.1073/pnas.0402638101
Hughes, H. V., Huitema, E., Pritchard, S., Keiler, K. C., Brun, Y. V, & Viollier, P. H. (2010). Protein localization and dynamics within a bacterial organelle. Proceedings of the National Academy of Sciences of the United States of America, 107(12), 5599–604. doi:10.1073/pnas.0909119107
Kwon, S.-K., Kim, B. K., Song, J. Y., Kwak, M.-J., Lee, C. H., Yoon, J.-H., … Kim, J. F. (2013). Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome biology and evolution, 5(1), 187–99. doi:10.1093/gbe/evs134
Lanyi, J. K., Duschl, A., Hatfield, G. W., May, K., & Wesley, G. (2013). The Primary Structure of a Halorhodopsin Natronobacterium pharaonis from.
Marks, M. E., Castro-Rojas, C. M., Teiling, C., Du, L., Kapatral, V., Walunas, T. L., & Crosson, S. (2010). The genetic basis of laboratory adaptation in Caulobacter crescentus. Journal of bacteriology, 192(14), 3678–88. doi:10.1128/JB.00255-10
McCusker, E. C., Bagnéris, C., Naylor, C. E., Cole, A. R., D’Avanzo, N., Nichols, C. G., & Wallace, B. a. (2012). Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nature communications, 3, 1102. doi:10.1038/ncomms2077
Ng, W. V, Kennedy, S. P., Mahairas, G. G., Berquist, B., Pan, M., Shukla, H. D., … DasSarma, S. (2000, October 24). Genome sequence of Halobacterium species NRC-1. Proceedings of the National Academy of Sciences of the United States of America. doi:10.1073/pnas.190337797
Obuchowski, P. L., & Jacobs-Wagner, C. (2008). PflI, a protein involved in flagellar positioning in Caulobacter crescentus. Journal of bacteriology, 190(5), 1718–29. doi:10.1128/JB.01706-07
Ohta, N., Lane, T., Ninfa, E. G., Sommer, J. M., & Newton, a. (1992). A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proceedings of the National Academy of Sciences of the United States of America, 89(21), 10297–301. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=50325&tool=pmcentrez&rendertype=abstract
Rüdiger, M., & Oesterhelt, D. (1997). Specific arginine and threonine residues control anion binding and transport in the light-driven chloride pump halorhodopsin. The EMBO journal, 16(13), 3813–21. doi:10.1093/emboj/16.13.3813
Thanbichler, M., Iniesta, A. a, & Shapiro, L. (2007). A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic acids research, 35(20), e137. doi:10.1093/nar/gkm818
Sciochetti, Stephen A., et al. "Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus." Journal of bacteriology 184.21 (2002): 6037-6049.
Zhang, F., Vierock, J., Yizhar, O., Fenno, L. E., Tsunoda, S., Kianianmomeni, A., … Deisseroth, K. (2011). The microbial opsin family of optogenetic tools. Cell, 147(7), 1446–57. doi:10.1016/j.cell.2011.12.004
 "
"