Team:UChicago/Notebook
From 2013.igem.org
(Difference between revisions)
(→Week 3) |
(→Week 4) |
||
| Line 32: | Line 32: | ||
===Week 4=== | ===Week 4=== | ||
| + | |||
| + | ====Wednesday, June 26, 2013==== | ||
| + | ---- | ||
| + | |||
| + | '''Planning Wet Lab Work''' | ||
| + | |||
| + | *We split into planning sub-teams to plan out different parts of the project with protocols, timelines, and ordering reagents. | ||
| + | **Plasmid Design | ||
| + | **Media and Competent Cells | ||
| + | **E. coli and B. subtilis Transformation | ||
| + | **Keratinase Assay and His tagging | ||
| + | **Running DNA gels, SDS-PAGE gels, and western blots | ||
| + | *We will also be holding weekly meetings with our graduate student advisers and biweekly meetings with our faculty advisers to provide updates. | ||
| + | *Wet lab work begins tomorrow! | ||
| + | |||
| + | |||
| + | ====Friday, June 28, 2013==== | ||
| + | ---- | ||
| + | |||
| + | '''Researching the Sequence of kerA''' | ||
| + | |||
| + | *Planning subteams continued to order reagents and compile protocols. | ||
| + | *The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT. | ||
| + | *kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995). | ||
| + | |||
| + | [[File:UChicago 2013 kerA plasmid design.png]] | ||
| + | |||
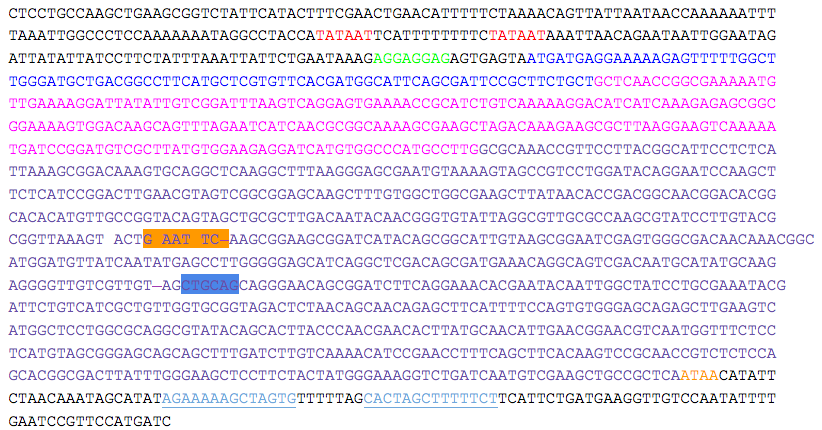
| + | *Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated. | ||
| + | *EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick. | ||
==July== | ==July== | ||
Revision as of 08:29, 26 September 2013
| Protocols |
|---|
Contents |
June
Week 3
Tuesday, June 18, 2013
First iGEM Meeting of the Summer!
- All student members gathered in the lab.
- We toured the lab, prep room, and incubator room, and we discussed the lab rules.
- We also split into teams to discuss lab logistics: contacting funding sources, ordering reagents/materials, compiling protocols, recording our electronic lab notebook, and making plates/media.
Week 4
Wednesday, June 26, 2013
Planning Wet Lab Work
- We split into planning sub-teams to plan out different parts of the project with protocols, timelines, and ordering reagents.
- Plasmid Design
- Media and Competent Cells
- E. coli and B. subtilis Transformation
- Keratinase Assay and His tagging
- Running DNA gels, SDS-PAGE gels, and western blots
- We will also be holding weekly meetings with our graduate student advisers and biweekly meetings with our faculty advisers to provide updates.
- Wet lab work begins tomorrow!
Friday, June 28, 2013
Researching the Sequence of kerA
- Planning subteams continued to order reagents and compile protocols.
- The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT.
- kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995).
- Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated.
- EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick.
July
Week 1
Week 2
Week 3
Week 4
August
Week 1
Week 2
Week 3
Week 4
September
Week 1
Week 2
Week 3
 "
"