Team:UChicago/Notebook
From 2013.igem.org
| Protocols |
|---|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Lab Notebook
Contents |
June
Week 3
Tuesday, June 18, 2013
First iGEM Meeting of the Summer!
- All student members gathered in the lab.
- We toured the lab, prep room, and incubator room, and we discussed the lab rules.
- We also split into teams to discuss lab logistics: contacting funding sources, ordering reagents/materials, compiling protocols, recording our electronic lab notebook, and making plates/media.
Week 4
Wednesday, June 26, 2013
Planning Wet Lab Work
- We split into planning sub-teams to plan out different parts of the project with protocols, timelines, and ordering reagents.
- Plasmid Design
- Media and Competent Cells
- E. coli and B. subtilis Transformation
- Keratinase Assay and His tagging
- Running DNA gels, SDS-PAGE gels, and western blots
- We will also be holding weekly meetings with our graduate student advisers and biweekly meetings with our faculty advisers to provide updates.
- Wet lab work begins tomorrow!
Friday, June 28, 2013
Researching the Sequence of kerA
- Planning subteams continued to order reagents and compile protocols.
- The plasmid design team decided on gibson assembly to produce the kerA biobrick with geneblocks ordered from IDT.
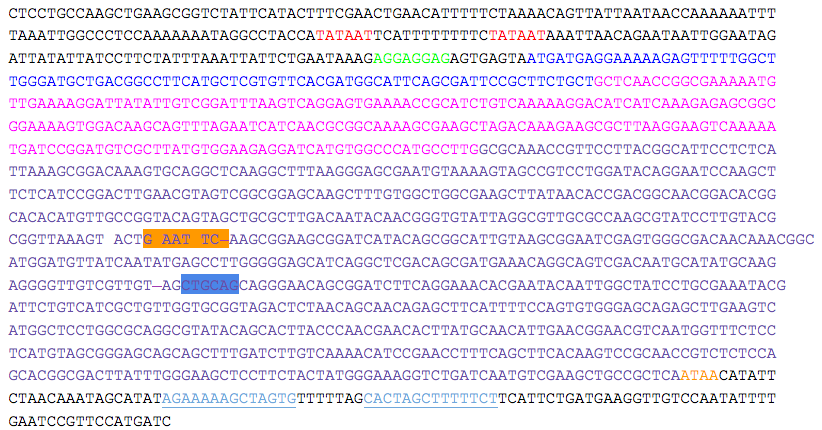
- kerA sequence was obtained from “Nucleotide Sequence and Expression of kerA, the Gene Encoding a Keratinolytic Protease of Bacillus licheniformis PWD-1” (Lin et al. 1995).
- Two putative promoters (red), a possible ribosomal binding site (green), and a transcriptional terminator sequence (light blue, single underline) are indicated. The putative starting residues of the preprotein (pre = blue), proprotein (pro = pink), and mature protein (mature = purple) are indicated.
- EcoRI (orange) and PstI (blue) sites within the gene are indicated and must be removed to produce the kerA biobrick.
July
Week 1
Wednesday, July 3, 2013
BioBrick Design Presentation and Grad Adviser Meeting
- Designed a biobrick with:
- C terminus His-tagged kerA
- CATCATCATCATCATCAT
- Prefix: gaattcgcggccgcttctagag
- Suffix: tactagtagcggccgctgcag
- C terminus His-tagged kerA
- Moreover, the bio brick must include
- Eliminated EcoRI and PstI restriction sites from kerA nucleotide seq
- EcoRI (gaattc--814) and PstI (ctgcag--978)
- Conserved aa seq + biobrick prefix + suffix
- Secretion signal peptide
- Inducible promoter
- Eliminated EcoRI and PstI restriction sites from kerA nucleotide seq
- We also presented a feasible timeline of our project, which included:
- Assembly of biobrick and transformation into E. coli
- Transformation of B. subtilis
- Keratinase isolation and keratinase assays
- kerA mutagenesis for optimization of the keratinase
- We also split up basic lab duties!
- Ivan & Ethan are in charge of the IBC protocol.
- We can’t start wet lab work until our IBC protocol is finished!
- Ethan is in charge of minutes.
- Susan is in charge of ordering invoices.
- Annie is in charge of emailing advisors and members to plan meetings
- Chloe is getting the B. subtilis gene that expressing keratinase.
- Ivan & Ethan are in charge of the IBC protocol.
Thursday, July 4, 2013
Milk Agar Plates
- The purpose of milk agar plates is often to detect protease activity, including the activity of keratinases.
- A measurement of any baseline protease activity of our parent B. subtilis strain (WB700), which does not endogenously express any keratinases, is desired.
- Excess protease activity of our parent B. subtilis strain may prevent our use of milk agar plates as an assay for keratinase expression of our future transformants with the kerA gene.
- Therefore, the parent B. subitilis strain (WB700) was streaked on the milk agar plates to observe baseline protease activity that could prevent us from detecting keratinase expression after transformation.
- [Milk Agar Plates] (in LB):
- Ideally, there should be a minimal zone of clearing around our B. subtilis colonies.
- Next Steps:
- Observe the results of our milk agar plates with B. subtilis colonies
Friday, July 5, 2013
Primer Design for kerA biobrick
- Designed primers for directed mutagenesis of our kerA biobrick using PrimerX:
- Primer pair 1 (EcoRI mutation, aat --> aac, asparagine)
- a. Forward: 5' GGTTAAAGTACTGAACTCAAGCGGAAGCGG 3'
- b. Reverse: 5' CCGCTTCCGCTTGAGTTCAGTACTTTAACC 3'
- Primer pair 2 (Pst1 mutation, gca-->gcg, alanine)
- a. Forward: 5' GTTGTCGTTGTAGCTGCGGCAGGGAACAGCGGATC 3'
- b. Reverse: 5' GATCCGCTGTTCCCTGCCGCAGCTACAACGACAAC 3'
- Used APE for checking codons and mutating restriction sites
Saturday, July 6, 2013
Primer Design Part II
- Designed primers to isolate non-coding and coding sequence of kerA and add prefix and suffix
- forward primer + xbaI site:
- 5' TATCTAGAGGTCTATTCATACTTTCGAA 3'
- reverse primer + speI site (reverse complement):
- 5' ATTGATCATTGGACAACCTTCATCAG 3'
- We still need to design additional primers that will include the secretion signal peptide (in case it needs to be secreted out of B. subtilis) and kerA (From start to stop codon).
- These primers should include igem biobrick prefix and his tag (before stop codon) + suffix.
Week 2
Monday, July 8, 2013
Milk Agar Plate Results
- The results of our milk agar assay from 7/04/13 of the non-transformed B. subtilis strain WB700:
- The zone of clearing around the B. subtilis colonies indicates that our parent strain breaks down milk. Its prominent endogenous protease activity prevents us from using the milk agar assay as a screen for successfully transformed kerA-expressing B. subtilis.
- Alternative method to screen for transformed B. subtilis needed:
- The feather assay used in Saha and Dhanasekaran’s 2010 paper, “Isolation and Screening of Keratinolytic Actinobacteria form Keratin Waste Dumped Soil in Tiruchirappalli and Nammakkal, Tamil Nadu, India”
- Quantitative assay using azokeratin
Our future plans:
- For our promoter for kerA, Karyl, one of our graduate advisors, recommended P43, a constitutive promoter, to transform into B. subtilis.
- We also decided to use pUB110, a high copy number plasmid used in S. aureus and B. subtilis, to introduce kerA into our parent B. subtilis strain.
- pUB110 will also be made into a biobrick.
- We have to choose a suitable plasmid (either pSB1A3, pSB1C3, pSB1T3, or pSB1K3.m1) for our E. coli transformations.
Wednesday, July 10, 2013
Grad Advisor Meeting on further developed Keratinase Project
- We presented the primers we designed to our advisers, who suggested we double check them for self dimerization and 3’ overlaps with:
- We discovered that our forward primer forms a self-dimer and must be redesigned.
- We also need lysozyme to digest the peptidoglycan of B. subtilis in order to miniprep it.
- We decided to use the iGEM plasmid pSB1K3.m1 for our E. coli transformations.
- To screen for keratinase activity, instead of using the feather assay, which takes up to a month for visible results to appear, we decided to use the azokeratin assay as the secondary screening method.
Week 3
Week 4
August
Week 1
Week 2
Week 3
Tuesday, August 20, 2013
kerA-pSB1A3 ligation and RFP digestion results
- Measured the concentration of the RFP digested yesterday
- [8/19 RFP Digest] = 74.7 ug/ul
- Ran a 2% gel of the RFP digest
- Lane 2: Ladder (8 ul)
- Lane 4: 8/19 RFP mp#2 Digest (2 ul) + loading dye (2 ul)
- Added 5/10 ul EtBr to the buffer
INSERT 8/20 GEL PIC HERE
- There is only 1 band in lane 4.
- Email iGEM to request new Pveg+RBS construct purchased
- Ligated kerA and pSB1A3 with control experiments
- The ligations were set up as follows:
Week 4
September
Week 1
Week 2
Week 3
 "
"