|
|
| (2 intermediate revisions not shown) |
| Line 7: |
Line 7: |
| | | | |
| | <p> | | <p> |
| - | To realize our idea of exploring and demonstrating the high biotechnological potential of the magnetosome nanoparticles, we had to find a way to execute this idea in a limited time frame and in a way that would get us results which also needed to be easily communicable to a wide audience. Our solution was linking a fluorescent protein to the magnetosome membrane. This fluorescent protein would give us results that could be communicated with just a simple picture of our bacterial cells and could be just as easily understood by everyone. In addition it would highlight one of the big advantages magnetosomes have over other nanomagnets, they are surrounded by a lipid bilayer, a feature which we wanted to utilize. The fluorescent protein could be easily replaced by for example a drug, another protein or a dye, paving the way for a wide range of possible applications in the future.</p>
| + | Our idea was to explore and demonstrate the great potential of applying magnetosome nanoparticles in biotechnology or nanomedicine. We had to find a way to execute the idea that:</p> |
| | + | <ul> |
| | + | <li><p>would result in useful tools to be added to the magnetosome toolbox</p></li> |
| | + | <li><p>would be executable in a limited time frame</p></li> |
| | + | <li><p>would get us results that could be easily communicated to a wide audience</p></li> |
| | + | </ul> |
| | + | |
| | + | <p>Our solution was linking a fluorescent protein to the magnetosome membrane. This fluorescent protein would give us results that could be communicated with just a simple picture of our bacterial cells and could be just as easily understood by everyone. Finally, it would highlight one of the big advantages that magnetosomes have over other nanomagnets - they are surrounded by a lipid bilayer, a feature which we wanted to utilize. The fluorescent protein could be easily replaced by for example a drug, another protein or a dye, paving the way for a wide range of <a href="https://2013.igem.org/Team:UNIK_Copenhagen/Project/Applications">exciting applications</a> in the future.</p> |
| | | | |
| | <div class="image"> | | <div class="image"> |
| | <a href="https://static.igem.org/mediawiki/2013/5/5e/UNIK_Copenhagen_Mgryphiswaldense.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/5/5e/UNIK_Copenhagen_Mgryphiswaldense.jpg"></a> | | <a href="https://static.igem.org/mediawiki/2013/5/5e/UNIK_Copenhagen_Mgryphiswaldense.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/5/5e/UNIK_Copenhagen_Mgryphiswaldense.jpg"></a> |
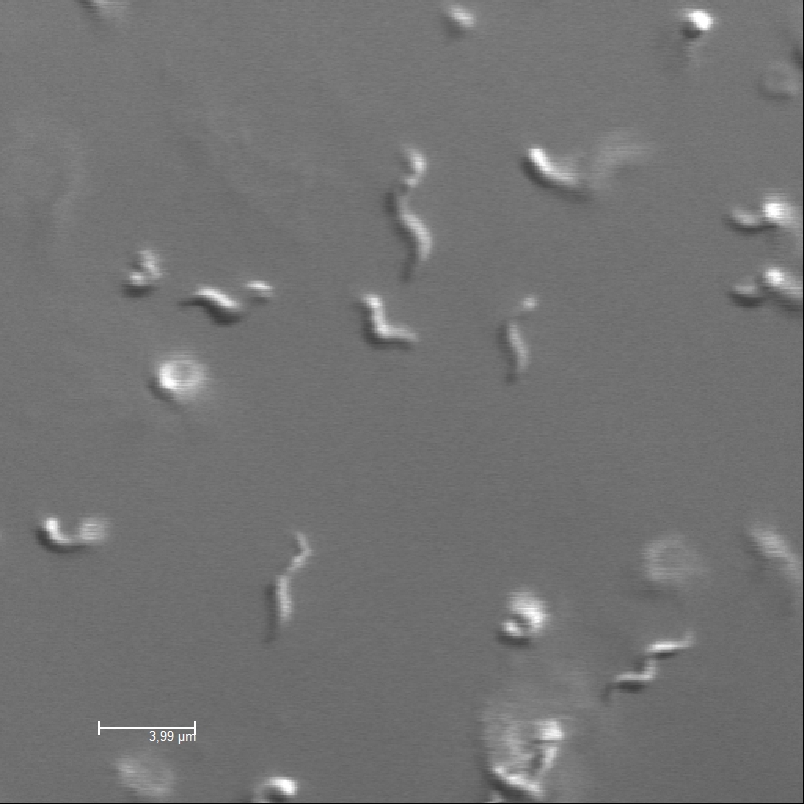
| - | <p>Confocal image of Magnetospirillum Gryphiswaldense</p> | + | <p>Confocal microscopy image of the magnetotactic bacterium <i>Magnetospirillum gryphiswaldense</i></p> |
| | </div> | | </div> |
| - | <b> MamC as a membrane anchor </b> | + | <p><b> MamC as a membrane anchor </b> <br> |
| - | <p>As a membrane anchor to link our fluorescent protein to the magnetosome we chose a protein called MamC that convinced us with its excellent properties. MamC is known for being highly abundant, very stable and non-interfering with the magnetosome formation itself which made it the perfect match for our experiment (Grünberg et al 2004; Scheffel et al. 2006). As our model organisms to work with in the lab, we chose two strains of magnetotactic bacteria that had also been used by the majority of researchers working in this field. Both <i>Magnetospirillum Gryphiswaldense</i> and <i>Magnetospirillum magnetotacticum</i> need anaerobic conditions to grow. For making our gene constructs of MamC plus fluorescent gene before moving them into the magnetotactic bacteria we decided to work with the well known all-rounder <i>Escherichia coli</i>. | + | As a membrane anchor to link our fluorescent protein to the magnetosome we chose a protein called MamC that convinced us with its excellent properties. MamC is known for being highly abundant, very stable and non-interfering with the magnetosome formation itself which made it the perfect match for our experiment (Grünberg et al 2004; Scheffel et al. 2006). As our model organisms to work with in the lab, we chose two strains of magnetotactic bacteria that had also been used by the majority of researchers working in this field. Both <i>Magnetospirillum gryphiswaldense</i> and <i>Magnetospirillum magnetotacticum</i> require anaerobic conditions to grow. For making our gene constructs of <i>mamC</i> plus a fluorescent protein gene to be transfected into the magnetotactic bacteria, we decided to work with the well-known model bacterium <i>Escherichia coli</i>. |
| | <br><br> | | <br><br> |
| - | <b> Fluorescent proteins GFP and FbFP </b> | + | <b> Fluorescent proteins GFP and FbFP </b> <br> |
| - | We then realized that we had to choose an additional fluorescent protein to work with other than the usually utilized green fluorescent protein (GFP). GFP shows green fluorescence when exposed to blue light. Since GFP requires oxygen for its chromophore production, which is responsible for its fluorescence, it could only be used efficiently with our aerobic strain. Therefore we chose a flavin mononucleotide–based fluorescent protein (FbFP), which is able to fluoresce without oxygen to use in our anaerobic strain (Drepper et al. 2007). | + | We then realized that we had to choose an additional fluorescent protein to work with, other than the usually utilized green fluorescent protein (GFP). GFP shows green fluorescence when exposed to blue light. Since GFP requires oxygen for its chromophore production, which is responsible for its fluorescence, it could only be used efficiently with our aerobic strain. Therefore we chose a flavin mononucleotide–based fluorescent protein (FbFP), which is able to fluoresce without oxygen to use in our anaerobic strain (Drepper et al. 2007). |
| | <br><br> | | <br><br> |
| - | <b> Isolation of local strains </b> | + | <b> Isolation of local strains </b> <br> |
| - | We also isolated our own Copenhagen strains of magnetotactic bacteria from a lake in Copenhagen. Further more, we got to sequence its genome and compare it to the genome of other magnetotactic bacteria. | + | We also isolated our own "Copenhagen" strains of magnetotactic bacteria from a lake in Copenhagen. Further more, we got to sequence its genome and compare it to the genome of other magnetotactic bacteria. |
| | </p> | | </p> |
| | | | |
| | <div class="image"> | | <div class="image"> |
| | <img src="https://static.igem.org/mediawiki/2013/f/f6/UNIK_Copenhagen_Experimental_MamC.jpg"> | | <img src="https://static.igem.org/mediawiki/2013/f/f6/UNIK_Copenhagen_Experimental_MamC.jpg"> |
| - | <p>Stylized magnetotactic bacterium, where you can see the magnetosomes with the MamC protein inside its membrane</p> | + | <p>A stylized magnetotactic bacterium, where you can see the magnetosomes with the MamC protein inside its membrane</p> |
| | </div> | | </div> |
| | | | |
| | <p> | | <p> |
| - | Going into the lab our strategy then was to isolate the MamC magnetosome gene from both our magnetotactic bacteria and fuse it to one of the fluorescent protein genes in a vector. | + | Going into the lab our strategy then was to isolate the MamC magnetosome gene from both our magnetotactic bacteria strains, <i>Magnetospirillum gryphiswaldense</i> and <i>Magnetospirillum magnetotacticum</i>, and fuse it to one of the fluorescent protein genes in a vector. |
| | </p> | | </p> |
| | | | |
| | <div class="image"> | | <div class="image"> |
| | <img src="https://static.igem.org/mediawiki/2013/7/7f/UNIK_Copenhagen_fullvector.jpg"> | | <img src="https://static.igem.org/mediawiki/2013/7/7f/UNIK_Copenhagen_fullvector.jpg"> |
| - | <p>The isolated MamC and fluorescent protein genes (GFP/FBFP) are cloned together into a vector</p> | + | <p>The isolated <i>mamC</i> and fluorescent protein genes (<i>gfp</i>/<i>fbfp</i>) are cloned together into a vector</p> |
| | </div> | | </div> |
| | | | |
| | <p> | | <p> |
| - | This vector serves as a plasmid that we can transfer into our magnetotactic bacteria, which will then express our MamC protein. MamC will be fluorescently labeled so that we can make it visible under blue light in a microscope. | + | This vector serves as a plasmid that we can transfer into our magnetotactic bacteria, which will then express the MamC protein. MamC will be fluorescently labeled so that we can make it visible under blue light in a microscope. |
| | </p> | | </p> |
| | | | |
| | <div class="image"> | | <div class="image"> |
| | <img src="https://static.igem.org/mediawiki/2013/1/1b/UNIK_Copenhagen_Experimental_MamC-GFP.jpg"> | | <img src="https://static.igem.org/mediawiki/2013/1/1b/UNIK_Copenhagen_Experimental_MamC-GFP.jpg"> |
| - | <p>Here you can see the MamC protein fused with one of our fluorescent proteins, the fluorescence it emits under blue light will be visible to the bare eye</p> | + | <p>A stylized illustration of the MamC protein fused with one of our fluorescent proteins, the fluorescence it emits under blue light will be visible to the naked eye</p> |
| | </div> | | </div> |
| | | | |
| | <p> | | <p> |
| - | Finally, when we make our bacteria express the MamC-flourescent protein construct in its magnetosomes, we can isolate them and further on study their behavior concerning future applications. | + | Finally, when our bacteria express the MamC-flourescent protein construct in its magnetosomes, we can isolate them and further on study their behavior concerning future applications. |
| | <br> | | <br> |
| | <br> | | <br> |



 "
"