|
|
| Line 8: |
Line 8: |
| | <a class="paneLink" href=".indepth">In-Depth</a> | | <a class="paneLink" href=".indepth">In-Depth</a> |
| | <a class="paneLink" href=".data">Data</a> | | <a class="paneLink" href=".data">Data</a> |
| - | <a class="paneLink" href=".modelling">Modeling</a> | + | <a class="paneLink" href=".references">References</a> |
| | </div> | | </div> |
| | | | |
| Line 72: |
Line 72: |
| | </div> | | </div> |
| | | | |
| - | <div class="row pane modelling"> | + | <div class="row pane references"> |
| - | <div class="zebra"> | + | <h1>References</h1> |
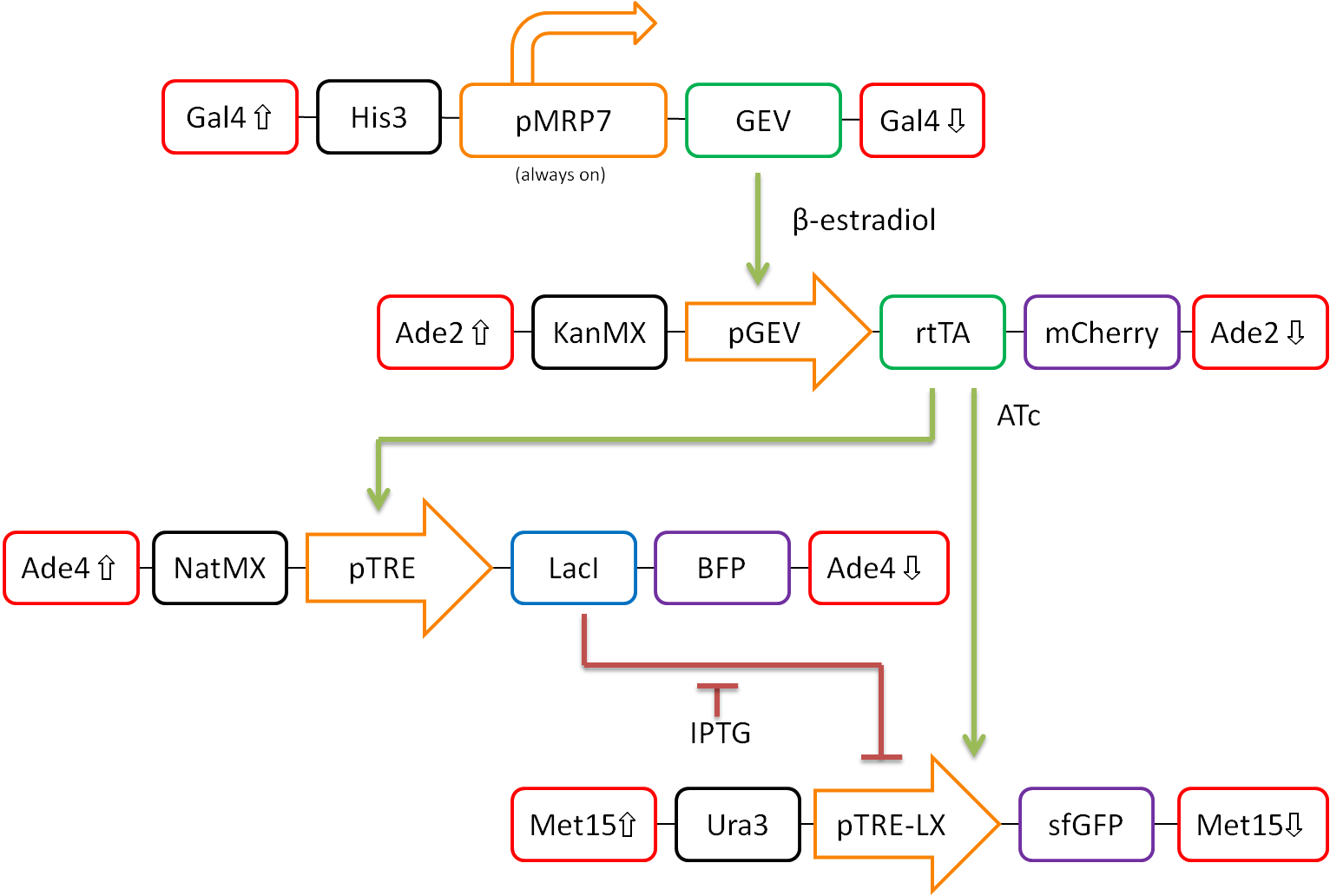
| - | <h2>Modeling the Type-I Incoherent Feedforward Loop</h2>
| + | <p>Under construction.</p> |
| - | <p style="color:black;">The Feedforward loop can be broken down into 3 main steps</p>
| + | </div> |
| - | <ol> | + | |
| - | <p style="margin-left:5em;">1. Toxin will stimulate production of X (rtTA).<p>
| + | |
| - | <p style="margin-left:5em;">2. X (rtTA) stimulates production of both Z (sfGFP) and Y (LacI).<p>
| + | |
| - | <p style="margin-left:5em;">3. Sufficient repression from Y terminates Z (sfGFP) production.<p>
| + | |
| - | </ol> | + | |
| - | <p>You can take a better look at the system by clicking on the button below!
| + | |
| - | <div id="mainContainer"style="height:200px; width:750px; margin-left:auto; margin-right:auto; vertical-align:middle; position:relative; top:50px;">
| + | |
| - | | + | |
| - | <img id="one"src="https://static.igem.org/mediawiki/2013/9/9c/One.png" height="75" width="75" style="margin-left:127.5px;"></img> | + | |
| - | <button id="start" onClick="toxMore();" style ="height:75px; width:350px; left:215px; background-color:white; border: 2px solid #07c; position:absolute; font-size:30px;cursor:pointer;">Click to add toxin!</button>
| + | |
| - | <br>
| + | |
| - |
| + | |
| - | <div id="toxinContainer" style=" height:200px; width:200px; margin-right:auto; margin-left:auto;">
| + | |
| - | <img id="toxin" src="https://static.igem.org/mediawiki/2013/e/eb/Tox.png" height="200" width="200" style="position:absolute;"></img>
| + | |
| - | <img id="toxinM" src="https://static.igem.org/mediawiki/2013/e/e2/ToxMore.png" height="200" width="200" style="position:absolute; display:none;"></img>
| + | |
| - | <canvas id="graph" height="330" width="350" style="position:absolute; top:300px; left:-110px;"></canvas>
| + | |
| - | </div>
| + | |
| - |
| + | |
| - | <div style="position:relative;height:500px; width:800px; margin-left:auto; margin-right:auto; top:-40px; ">
| + | |
| - | | + | |
| - | <p id="equation" style="font-size:45px; top:-70px; left:585px; position:absolute; display:none;">Equations</p>
| + | |
| - | <p id="graphId" style="font-size:45px; top:-70px; left:-65px; position:absolute; display:none;">Graph</p>
| + | |
| - | <img id="bigArrow" src="https://static.igem.org/mediawiki/2013/8/85/BigArrow.png" height="350" width="93" style="position: absolute; top: 60px; left: 240px; display:none;"></img>
| + | |
| - | | + | |
| - | <p id = "theX" align = "center" style="font-size:80px; left:325px; display:none; overflow:hidden; position:absolute; color:blue; " > X </p>
| + | |
| - | | + | |
| - | <p id = "titledX" align = "center" style="font-size:15px; left:395px; top:28px; display:none; overflow:hidden; position:absolute;" > 1. Variation of X over variation of time </p>
| + | |
| - | | + | |
| - | <p id = "titledY" align = "center" style="font-size:15px; left:395px; top:200px; display:none; overflow:hidden; position:absolute;" > 4. Variation of Y over variation of time </p>
| + | |
| - | | + | |
| - | <p id = "titledZ" align = "center" style="font-size:15px; left:395px; top:350px; display:none; overflow:hidden; position:absolute;" > 5. Variation of Z over variation of time </p>
| + | |
| - | | + | |
| - | <img id="xEquation" src="https://static.igem.org/mediawiki/2013/a/ad/EquationX.png" height="41" width="460" style="position: absolute; top: 50px; left: 390px; display:none;"></img>
| + | |
| - |
| + | |
| - | <img id="yEquation" src="https://static.igem.org/mediawiki/2013/6/65/EquationY.png" height="41" width="460" style="position: absolute; top: 220px; left: 390px; display:none;"></img>
| + | |
| - | | + | |
| - | <img id="zEquation" src="https://static.igem.org/mediawiki/2013/3/3d/EquationZ.png" height="41" width="460" style="position: absolute; top: 370px; left: 390px; display:none;"></img>
| + | |
| - |
| + | |
| - | <img id="downArrow" src="https://static.igem.org/mediawiki/2013/a/af/DownArrowUottawa.png" height="120" width="22" style="position:absolute;left:335px; top: 100px; display:none;"></img>
| + | |
| - | | + | |
| - | <img id="affinityX" src="https://static.igem.org/mediawiki/2013/c/ca/ActiveX.png" height="50" width="265" style="position: absolute; top: 150px; left: 350px; display:none;"></img>
| + | |
| - | | + | |
| - | <img id="affinityY" src="https://static.igem.org/mediawiki/2013/6/66/ActiveY.png" height="50" width="265" style="position: absolute; top: 300px; left: 350px; display:none;"></img>
| + | |
| - | | + | |
| - | <p id = "titledAY" align = "center" style="font-size:15px; left:350px; top:280px; display:none; overflow:hidden; position:absolute;" > 3. Amount of Active Y relative to amount of IPTG</p>
| + | |
| - | <p id = "titledAX" align = "center" style="font-size:15px; left:350px; top:120px; display:none; overflow:hidden; position:absolute;" > 2. Amount of Active X relative to amount of ATC</p>
| + | |
| - | | + | |
| - | <p id = "theY" align = "center" style="font-size:80px; position:absolute; left:325px; top:170px; opacity:0.2; display:none; color:green; "> Y </p>
| + | |
| - |
| + | |
| - | <img id="repressionArrow" src="https://static.igem.org/mediawiki/2013/b/b7/RepressionArrow.png" height="120" width="22" style="position: absolute; top: 270px; left: 335px; display:none; "></img>
| + | |
| - | | + | |
| - | <p id = "theZ" align = "center" style="font-size:80px; top:340px; position:absolute; left:335px; opacity:0.2; color:red; display:none;"> Z </p>
| + | |
| - |
| + | |
| - | <img id="two" src="https://static.igem.org/mediawiki/2013/c/cd/Two.png" height="75" width="75" style="top:450px; margin-left:-47.5px; position:absolute; display:none; overflow:hidden;"></img>
| + | |
| - | <button id="spikeZ" onClick="fadeZs();drawZ();" style ="top:450px; height:75px; width:320px; margin-left:47.5px; background-color:white; border: 2px solid #FF0000; position:absolute; font-size:30px; position:absolute; cursor:pointer; display:none; overflow:hidden;">Click to show activation by X!</button>
| + | |
| - | | + | |
| - | <img id="three" src="https://static.igem.org/mediawiki/2013/f/f4/Three.png" height="75" width="75" style="top:450px; margin-left:400px; position:absolute; display:none; overflow:hidden;"></img>
| + | |
| - | <button id="spikeY" onClick="fadeYs();drawY()" style ="top:450px; height:75px; width:350px; margin-left:480px; background-color:white; border:2px solid green; position:absolute; font-size:30px; position:absolute; cursor:pointer; display:none; overflow:hidden; " align="right"> Click to show repression by Y!</button>
| + | |
| - | <button id="restartButton" onClick="document.location.reload(true)" style="top:460px; height:85px; width:300px; margin-left:200px; display:none; position:absolute; font-size:25px; background-color:white; border:2px solid blue; cursor:pointer;">Restart!</button>
| + | |
| - | </div>
| + | |
| - | | + | |
| - | </div>
| + | |
| - | | + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | <h2>Equations involved in modeling the biological system</h2>
| + | |
| - | <br>
| + | |
| - | <p> The mathematical model was developed using basic rate formulas of activation and repression as shown in the system of equations above. We then expanded them to include the different parameters required for our model.</p>
| + | |
| - | <p>These additions are as follows</p>
| + | |
| - | <p style="margin-left:5em">IPTG concentration to calculate the amount of Y (LacI) repressing Z (sfGFP).</p>
| + | |
| - | <p style="margin-left:5em">ATC concentration to calculate the amount of X (rtTA) available to bind Y promoter and Z promoter.</p>
| + | |
| - | <p style="margin-left:5em">Transfer rates of proteins between the cytosol and the nuclei to calculate transfer rates by size of each protein.</p>
| + | |
| - | <br>
| + | |
| - | <p>The four rate equations for the production of mRNA over time of GEV (1.1), X (rtTA) (2.1), Y (LacI) (3.1) and Z (sfGFP) (4.1) in the nucleus are show below. </p>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <div style="padding-right:5%;padding-left:5%;margin-left:auto; margin-right:auto;"><img src="https://static.igem.org/mediawiki/2013/0/0c/Set1mRNA.png"></img></div>
| + | |
| - | | + | |
| - | <button onmousedown="slidedown('legendmRNA')"onclick="slideup('legendmRNA')" style="font-size:18px; font-family:Helvetica; maging-left:auto; margin-right:auto; width:auto; cursor:pointer;" align="left">Click for legend</button>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <div id="legendmRNA" style="height:300px;overflow:auto;width:auto;position:relative;display:none;margin-left:50px;font-size:12px;">
| + | |
| - | 1.1 Variation of GEV mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc"><b>MRP7</b> is the constitutive transcription rate that does not change. Replicates the role of a toxin in the system.</li>
| + | |
| - | <li type="disc"><b>Trs<sub>GEV mRNA</sub></b> is the ratio of the amount of GEV mRNA that will be transfered to the cytosol to be translated into protein. This value is set to 1.</li>
| + | |
| - | <li type="disc"><b>leakage</b> is the basic amount of GEV mRNA that is produced per second without activation</li>
| + | |
| - | </ol>
| + | |
| - | 2.1 Variation of rtTA mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc"><b>pGEV</b> is the fully induced transcription rate of this mRNA lenght.</li>
| + | |
| - | <li type="disc"><b>K<sub>GEV</sub></b> is the affinity of the GEV to the rtTA promoter.</li>
| + | |
| - | <li type="disc"><b>GEV<sub>Bound</sub></b> is the Amount of activated GEV by ß-estradiol from equation 3.0.</li>
| + | |
| - | <li type="disc"><b>n</b> is the cooperativity of the molecule to the promoter or the Hill coefficient.</li>
| + | |
| - | <li type="disc"><b>Trs<sub>rtTA mRNA</sub></b> is the ratio of the amount of rtTA mRNA that will be transfered to the cytosol to be translated into protein. This is set to 1.</li>
| + | |
| - | <li type="disc"><b>rtTA<sub>mRNA</sub></b> is the amount of rtTA mRNA transcribed by the promoter.</li>
| + | |
| - | <li type="disc"><b>leakage</b> is the basic amount of rtTA mRNA that is produced per second without activation.</li>
| + | |
| - | </ol>
| + | |
| - | 3.1 Variation of LacI mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc">Values in equation 3 have the same role as in equation 2.0. Only the molecule's names have been substituded.</li>
| + | |
| - | </ol>
| + | |
| - | 4.1 Variation of sfGFP mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc"><b>psfGFP</b> is the fully induced transcription rate of this mRNA lenght.</li>
| + | |
| - | <li type="disc"><b>K<sub>GEV</sub></b> is the affinity of the GEV to the rtTA promoter</li>
| + | |
| - | <li type="disc"><b>GEV<sub>Bound</sub></b> is the Amount of activated GEV by ß-estradiol</li>
| + | |
| - | <li type="disc"><b>n</b> is the cooperativity of the molecule to the promoter</li>
| + | |
| - | <li type="disc"><b>LacI<sub>free</sub></b> is the amount of LacI available to repress the activation sfGFP</li>
| + | |
| - | <li type="disc"><b>K<sub>LacI</sub></b> is the affinity of the LacI to the sfGFP promoter</li>
| + | |
| - | <li type="disc"><b>Trs<sub>sfGFP mRNA</sub></b> transfer rate of the mRNA from the nucleus to the cytosol. This is set to 1.</li>
| + | |
| - | <li type="disc"><b>sfGFP<sub>mRNA</sub></b> amount of mRNA transcribed in the nucleus.</li>
| + | |
| - | <li type="disc"><b>leakage</b> is the basic amount of GEV mRNA that is produced per second without activation.</li>
| + | |
| - | </ol>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | </div>
| + | |
| - | <p>The first sets of equations were used to describe the amount of mRNA transcribed. These equations also took into consideration that the mRNA would be transported out of the nuclei. In the model, we assumed that all of the mRNA molecules would be exported to the cytosol. The differential equations representing the variation in mRNA in the cytosol are represented below in equations 1.2, 2.2, 3.2 and 4.2.</p>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <div style="padding-right:5%;padding-left:5%;margin-left:auto; margin-right:auto;"><img src="https://static.igem.org/mediawiki/2013/4/45/Set2mRNA.png"></img></div>
| + | |
| - | <br>
| + | |
| - | <button onmousedown="slidedown('legendmRNA2')"onclick="slideup('legendmRNA2')" style="font-size:18px; font-family:Helvetica; maging-left:auto; margin-right:auto; width:auto;" align="left">Click for legend</button>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <div id="legendmRNA2" style="height:300px;overflow:auto;width:auto;position:relative;display:none;margin-left:50px;font-size:12px;">
| + | |
| - | 1.2 Variation of GEV mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc"><b>Trs<sub>GEV mRNA</sub></b> is the ratio of the amount of GEV mRNA that was transfered to the cytosol. This value is set to 1.</li>
| + | |
| - | <li type="disc"><b>GEV<sub>mRNA</sub></b> is the amount of mRNA transcribed in the nucleus.</li>
| + | |
| - | <li type="disc"><b>Deg<sub>GEV mRNA</sub></b> the average degradation rate of an mRNA molecule in a cell.</li>
| + | |
| - | <li type="disc"><b>Dil</b> the rate at which the cell grows which is proportional to the rate at which the mRNA dillutes in the cell</li>
| + | |
| - | <li type="disc"><b>Cyt<sub>GEV mRNA</sub></b> amount of GEV mRNA in the cytosol.</li>
| + | |
| - | </ol>
| + | |
| - | 2.2 Variation of rtTA mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc">Values have same roles, but different names</li>
| + | |
| - | </ol>
| + | |
| - | 3.2 Variation of LacI mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc">Values have same roles, but different names</li>
| + | |
| - | </ol>
| + | |
| - | 4.2 Variation of sfGFP mRNA according to time.
| + | |
| - | <ol>
| + | |
| - | <li type="disc">Values have same roles, but different names</li>
| + | |
| - | </ol>
| + | |
| - | <br>
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <p>Computing the variation and amounts of mRNA in the cytosol over time then allowed us to compute the variation of protein translated. The equations of translation shown below in equations 1.3, 2.3, 3.3, 4.3 represent the variation of protein in the cytosol over time. The translational rates were calculated based on the average translation rate of a ribosome (9.5 base pair per second <sup>12</sup>). Here, we assume that the quantity of ribosome is not a limiting factor and that there are no translational regulations involved in the model. The degradation rates are taken from the average degradation rate of protein in yeast<sup>14</sup>.</p>
| + | |
| - | <div style="padding-right:10%; padding-left:10%;margin-left:auto; margin-right:auto;"><img src="https://static.igem.org/mediawiki/2013/9/92/ProteinCytosol.png"></img></div>
| + | |
| - | | + | |
| - | <button onmousedown="slidedown('legendProtein')"onclick="slideup('legendProtein')" style="font-size:18px; font-family:Helvetica; maging-left:auto; margin-right:auto; width:auto; cursor:pointer;" align="left">Click for legend</button>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <div id="legendProtein" style="height:300px;overflow:auto;width:auto;position:relative;display:none;margin-left:50px;font-size:12px;">
| + | |
| - | 1.3 Variation of GEV in the cytosol over time.
| + | |
| - | <br>
| + | |
| - | 2.3 Variation of rtTA in the cytosol over time.
| + | |
| - | <br>
| + | |
| - | 3.3 .Variation of LacI in the cytosol over time.
| + | |
| - | <br>
| + | |
| - | 4.3 Variation of Z (sfGFP) in the cytosol over time.
| + | |
| - | <br>
| + | |
| - | <b>Legend for variable that apply for all equations 4 equations</b>
| + | |
| - | <ol>
| + | |
| - | <li type="disc"><b>FullTrans</b> the full translation rate of the mRNA into protein. The rate is calculated based on the size of protein and vary for each protein</li>
| + | |
| - | <li type="disc"><b>Transf</b> transfer rate of the protein from the cytosol back into the nuclues. Extrapolated from known data and varies based on protein size. Equation 4.4 does not have this component because sfGFP only needs to be observed and does not have an effect on transcription of other genes.</li>
| + | |
| - | <li type="disc"><b>deg<sub>protein</sub></b> the average rate of degradation of protein. 3.5 molecules/hour in the cell.</li>
| + | |
| - | <li type="disc"><b>Dil</b> the rate at which the cell grows which is proportional to the rate at which the mRNA dillutes in the cell</li>
| + | |
| - | <li type="disc">The amounts of protein are represented by their name<sub>Cyt</sub></li>
| + | |
| - | </ol>
| + | |
| - | | + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <p>Once the amounts of protein calculated, we assumed that they would diffuse at an average rate through the nuclear pore to then affect the transcription of the other genes in the system. The calculations leading to the transfer rates are described in the parameter section under transf. Equations 1.4, 2.4, 3.4 and 4.4 represent the variation of protein present in the nucleus over time.</p>
| + | |
| - | | + | |
| - | <br>
| + | |
| - | | + | |
| - | <div style="padding-right:10%; padding-left:10%;margin-left:auto; margin-right:auto;"><img src="https://static.igem.org/mediawiki/2013/9/99/ProteinNucleus.png"></img></div>
| + | |
| - | <br>
| + | |
| - | <button onmousedown="slidedown('legendProteinN')"onclick="slideup('legendProteinN')" style="font-size:18px; font-family:Helvetica; maging-left:auto; margin-right:auto; width:auto; cursor:pointer;" align="left">Click for legend</button>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | <div id="legendProteinN" style="height:300px;overflow:auto;width:auto;position:relative;display:none;margin-left:50px;font-size:12px;">
| + | |
| - | 1.4 Variation of GEV in the nucleus over time.
| + | |
| - | <br>
| + | |
| - | 2.4 Variation of rtTA in the nucleus over time.
| + | |
| - | <br>
| + | |
| - | 3.4 .Variation of sfGFP in the nucleus over time.
| + | |
| - | <br>
| + | |
| - | <b>Legend for variable that apply for all equations 4 equations</b>
| + | |
| - | <ol>
| + | |
| - | <li type="disc"><b>Transf</b> transfer rate of the protein from the cytosol back into the nuclues. Extrapolated from known data and varies based on protein size.</li>
| + | |
| - | <li type="disc">The amounts of protein are represented by their name<sub>Cyt</sub></li>
| + | |
| - | <li type="disc"><b>deg<sub>protein</sub></b> the average rate of degradation of protein. 3.5 molecules/hour in the cell<sup>14</sup>.</li>
| + | |
| - | <li type="disc"><b>Dil</b> the rate at which the cell grows which is proportional to the rate at which the mRNA dillutes in the cell</li>
| + | |
| - | <li type="disc">The amounts of protein in the nuclues are represented by their name<sub>Nuc</sub></li>
| + | |
| - |
| + | |
| - | </ol>
| + | |
| - | | + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <p>To have better control on the feed-forward loop system, LacI was regulated with IPTG. Changing the amount of ATC regulated the amount of active rtTA. And the Amount of GEV available to activate production of rtTA is controlled with ß-estradiol. The following equations used to compute the amount of each compound are shown below. These numbers are imputed in the mRNA equations and vary accordingly with the amount of each transcription factors expressed in the ODE system.</p>
| + | |
| - | <div style="padding-left:5%; padding-right:5%; margin-left:auto; margin-right:auto;"><img src="https://static.igem.org/mediawiki/2013/6/6f/ControlEquations.png"></img></div
| + | |
| - | <br>
| + | |
| - | | + | |
| - | <button onmousedown="slidedown('legendActive')"onclick="slideup('legendActive')" style="font-size:18px; font-family:Helvetica; maging-left:auto; margin-right:auto; width:auto; cursor:pointer;" align="left">Click for legend</button>
| + | |
| - | <div id="legendActive" style="height:190px;width:auto;position:relative;overflow:auto;display:none;margin-left:50px;font-size:12px;">
| + | |
| - | <b>LacI<sub></sub>free</b> = the amount of LacI that is not bound to the IPTG. The LacI that is not bound to the IPTG is able to repress the expression of Z (sfGFP). Therefore the more IPTG present the more expression of Z is goign to be strong.
| + | |
| - | <br>
| + | |
| - | <b>rtTA<sub>bound</sub> </b>= the amount of rtTA that is available to activatite the Z (sfGFP) promoter and the Y (LacI) promoters. rtTA activity is regulated by ATC. The more ATC is present the stronger the expression of rtTA will be.
| + | |
| - | <br>
| + | |
| - | <b>GEV<sub>bound</sub> </b>= the amount of GEV that is available to activatite the X (rtTA) promoter. GEV activity is regulated by ß-estradiol. The more ß-estradiol present the stronger the expression of X (rtTA) will be.
| + | |
| - | <br>
| + | |
| - | <b>K</b> are the affinity constants of the different compounds.
| + | |
| - | </div>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <h2>Parameters</h2>
| + | |
| - | <p>Some parameters were found in the literature. Others were derived from other known constants as explained below.</p>
| + | |
| - | <p><b>Transcription rate</b><p>
| + | |
| - | <p>The transcription rate was calculated using the known rate of 2kb per minutes<sup>6</sup> which translates to 33.3 nucleotides per second. This rate was then divided by the amount of nucleotides in one mRNA strand. The values are in the parameter table.</p>
| + | |
| - | <p><b>Translation rate</b><p>
| + | |
| - | <p>The translation rate was calculated using the known rate of 9.5 amino acid per second<sup>12</sup>. This rate was then divided by the amount of amino acids per protein. The values are in the parameter table.</p>
| + | |
| - | <p><b>Transfer rate of protein from the cytosol to the nucleus</b><p>
| + | |
| - | <p>The rate of transfer from the cytosol to the nucleus was extrapolated from the formula rate = 0.01*(5000-size of protein in Dalton)-50. This is the multiplied by 119 or the average number of nuclear pores in yeast<sup>7</sup>. The first equation was determined by assuming that transfer rate of protein 5000 Da takes 0 seconds and a 17000 Da protein takes 120 seconds<sup>11</sup>.</p>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <table class=MsoTableGrid border=0 cellspacing=0 cellpadding=0
| + | |
| - | style='border-collapse:collapse;border:none;mso-yfti-tbllook:1184;mso-padding-alt:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt;mso-border-insideh:none'>
| + | |
| - | <tr style='mso-yfti-irow:0;mso-yfti-firstrow:yes'>
| + | |
| - | <td width=170 valign=top style='width:6.0cm;border-top:none;border-left:none;
| + | |
| - | border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;
| + | |
| - | padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-left-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>GEV</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-left-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span class=SpellE><span
| + | |
| - | class=GramE><span lang=EN-US>rtTA</span></span></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-left-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span class=SpellE><span
| + | |
| - | lang=EN-US>LacI</span></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-left-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span class=SpellE><span
| + | |
| - | class=GramE><span lang=EN-US>sfGFP</span></span></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-left-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>IPTG</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-left-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>ß-estradiol</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-left-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>ATC</span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | <tr style='mso-yfti-irow:1'>
| + | |
| - | <td width=170 style='width:6.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-top-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span lang=EN-US>Translation rates (mRNA/s)</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.02088</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.01503</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.01599</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.03991</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | <tr style='mso-yfti-irow:2'>
| + | |
| - | <td width=170 style='width:6.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-top-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span class=GramE><span lang=EN-US>n</span></span><span
| + | |
| - | lang=EN-US> (Hill coefficient)</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>1</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>2</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>2</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>-</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>2</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>1</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>1</span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | <tr style='mso-yfti-irow:3'>
| + | |
| - | <td width=170 style='width:6.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-top-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span lang=EN-US>K</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>10^-6<sup>13</sup></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>5*10^-4<sup>13</sup></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>10^-10 <sup>1</sup></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>-</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>5*10^-6<sup>3</sup></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>10^-9<sup>4</sup></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>1.56*10^-9<sup>13</sup></span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | <tr style='mso-yfti-irow:4'>
| + | |
| - | <td width=170 style='width:6.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-top-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span class=SpellE><span lang=EN-US>FullTrans</span></span><span
| + | |
| - | lang=EN-US> (molecules/second)</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.02442</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.01758</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.01870</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.04668</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | <tr style='mso-yfti-irow:5'>
| + | |
| - | <td width=170 style='width:6.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-top-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span class=SpellE><span lang=EN-US>Transf</span></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.297</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.19993</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>0.2150</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;mso-border-alt:
| + | |
| - | solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US><o:p> </o:p></span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | <tr style='mso-yfti-irow:6'>
| + | |
| - | <td width=170 style='width:6.0cm;border-top:none;border-left:none;border-bottom:
| + | |
| - | solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;mso-border-top-alt:
| + | |
| - | solid windowtext .5pt;mso-border-top-alt:solid windowtext .5pt;mso-border-bottom-alt:
| + | |
| - | solid windowtext .5pt;mso-border-right-alt:solid windowtext .5pt;padding:
| + | |
| - | 0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal><span lang=EN-US>Leakage</span></p>
| + | |
| - | </td>
| + | |
| - | <td width=85 colspan=7 style='width:3.0cm;border:none;border-bottom:solid windowtext 1.0pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-top-alt:solid windowtext .5pt;mso-border-left-alt:solid windowtext .5pt;
| + | |
| - | mso-border-bottom-alt:solid windowtext .5pt;padding:0cm 5.4pt 0cm 5.4pt'>
| + | |
| - | <p class=MsoNormal align=center style='text-align:center'><span lang=EN-US>1.5
| + | |
| - | molecule/hour for one promoter</span></p>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | <h2>References</h2>
| + | |
| - | <ol>
| + | |
| - | <li type="1" style="font-size:12px;"><a href="http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=100741&ver=0">http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=100741&ver=0</a> K of LacI to DNA.</li>
| + | |
| - | <li type="2" style="font-size:12px;"><a href="http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=100743&ver=0&trm=laci">http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=100743&ver=0&trm=laci</a> K of IPTG to LacI.</li>
| + | |
| - | <li type="3" style="font-size:12px;"><a href="http://pubs.acs.org/doi/pdf/10.1021/jp308930c">http://pubs.acs.org/doi/pdf/10.1021/jp308930c</a> K of IPTG to LacI.</li>
| + | |
| - | <li type="4" style="font-size:12px;"><a href="http://www.rcsb.org/pdb/explore/explore.do?structureId=1ORK">http://www.rcsb.org/pdb/explore/explore.do?structureId=1ORK</a></li>
| + | |
| - | <li type="5" style="font-size:12px;"><a href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC373162/">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC373162/</a> Time division of yeast</li>
| + | |
| - | <li type="6" style="font-size:12px;"><a href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3154325/pdf/nihms314191.pdf">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3154325/pdf/nihms314191.pdf</a> Fully induced transcription</li>
| + | |
| - | <li type="7" style="font-size:12px;"><a href="http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=101395&ver=1&trm=pore%20yeast">http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=101395&ver=1&trm=pore%20yeast</a> number of pores</li>
| + | |
| - | <li type="8" style="font-size:12px;"><a href="https://2009.igem.org/Team:PKU_Beijing/Modeling/ODE">https://2009.igem.org/Team:PKU_Beijing/Modeling/ODE</a> calculation of rate</li>
| + | |
| - | <li type="9" style="font-size:12px;"><a href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC373162/">http://www.ncbi.nlm.nih.gov/pmc/articles/PMC373162/</a> Time division of yeast</li>
| + | |
| - | <li type="10" style="font-size:12px;"><a href="http://www.nature.com/nbt/journal/v30/n7/pdf/nbt.2281.pdf">http://www.nature.com/nbt/journal/v30/n7/pdf/nbt.2281.pdf</a> Half life of sfGFP</li>
| + | |
| - | <li type="11" style="font-size:12px;"><a href="http://www.ncbi.nlm.nih.gov/books/NBK26932/">http://www.ncbi.nlm.nih.gov/books/NBK26932/</a> speed of pore</li>
| + | |
| - | <li type="12"style="font-size:12px;"><a href="http://download.cell.com/pdf/PIIS0092867413006557.pdf?intermediate=true">http://download.cell.com/pdf/PIIS0092867413006557.pdf?intermediate=true</a> Rate of translation</li>
| + | |
| - | | + | |
| - | <li type="13" style="font-size:12px;">Approximated to adapt the model</li>
| + | |
| - | <li type="14" style="font-size:12px;">Dmitry Nevozhaya, Rhys M. Adamsa, Kevin F. Murphyb, Kresˇimir Josic, and Ga bor Bala zsia, Negative autoregulation linearizes the dose–response and suppresses the heterogeneity of gene expression, <a href="www.pnas.org/cgi/doi/10.1073/pnas.0809901106">www.pnas.org/cgi/doi/10.1073/pnas.0809901106</a></li>
| + | |
| - | | + | |
| - | </ol>
| + | |
| - | | + | |
| | | | |
| | </div> | | </div> |
| | </div> | | </div> |
| | </html> | | </html> |




 "
"

