|
|
| Line 2: |
Line 2: |
| | {{:Team:ETH_Zurich/Templates/stylesheet}} | | {{:Team:ETH_Zurich/Templates/stylesheet}} |
| | | | |
| - | <h1>Cloned constructs</h1> | + | <h1>Final Circuit</h1> |
| | | | |
| - | <p>For the final Colisweeper circuit we plan a four plasmid system. The mine cells constitutively express LuxI and NagZ. In the Non-Mine cells LuxR and PhoA are expressed constitutively whereas Aes and GusA are expressed from pLux promoters with different sensitivities. To get to this system we tested different versions of the circuit with GFP. In the following table we list all the biobricks we used, the plasmids we cloned and what experiments we used them for. In general we used standard biobrick cloning techniques as described in the methods section. Whenever we used PCR gene amplification for cloning, we list the primers used in the following table. To be able to co-transform different plasmids we used backbones with compatible origins of replication and resistance genes. In the table you can find which backbone versions we used for which constructs.</p> | + | <p>For the final Colisweeper circuit we plan a four plasmid system. The mine cells constitutively express LuxI for signal generation and NagZ as identifier hydrolase. In the non-mine cells LuxR is expressed constitutively to process the OHHL signal. PhoA is expressed constitutively as well as reporter for safe cells. Aes and GusA are expressed from pLux promoters with different sensitivities. You can find all the biobricks we used and our own new biobricks in the figure below.</p> |
| | | | |
| | [[File:Plasmidmap.png|1050px|left|thumb|<b>Figure 1. Plasmids in mine and non-mine cells </b>]] | | [[File:Plasmidmap.png|1050px|left|thumb|<b>Figure 1. Plasmids in mine and non-mine cells </b>]] |
| | <br clear="all"/> | | <br clear="all"/> |
| | + | |
| | + | <br><h1>Cloned Constructs</h1> |
| | + | <p>To get to the circuit mentioned above we tested different versions of the circuit. For example we started our experiments using GFP as a reporter instead of the hydrolases. Then we also tested different LuxI and LuxR generating constructs. In the following table we list all the biobricks we used, the plasmids we cloned and what experiments we used them for. In general we used standard biobrick cloning techniques as described in the methods section. Whenever we used PCR gene amplification for cloning, we list the primers used in the following table. To be able to co-transform different plasmids we used backbones with compatible origins of replication and resistance genes. In the table you can find which backbone versions we used for which constructs.</p> |
| | | | |
| | <table style="float:left;margin-top:10px;width:auto;height:auto;font-size:12px"> | | <table style="float:left;margin-top:10px;width:auto;height:auto;font-size:12px"> |
Final Circuit
For the final Colisweeper circuit we plan a four plasmid system. The mine cells constitutively express LuxI for signal generation and NagZ as identifier hydrolase. In the non-mine cells LuxR is expressed constitutively to process the OHHL signal. PhoA is expressed constitutively as well as reporter for safe cells. Aes and GusA are expressed from pLux promoters with different sensitivities. You can find all the biobricks we used and our own new biobricks in the figure below.

Figure 1. Plasmids in mine and non-mine cells
Cloned Constructs
To get to the circuit mentioned above we tested different versions of the circuit. For example we started our experiments using GFP as a reporter instead of the hydrolases. Then we also tested different LuxI and LuxR generating constructs. In the following table we list all the biobricks we used, the plasmids we cloned and what experiments we used them for. In general we used standard biobrick cloning techniques as described in the methods section. Whenever we used PCR gene amplification for cloning, we list the primers used in the following table. To be able to co-transform different plasmids we used backbones with compatible origins of replication and resistance genes. In the table you can find which backbone versions we used for which constructs.
| GFP constructs |
| |
Description |
Cloning |
Maps |
| 1 |
Receiver cell construct for GFP diffusion experiments |
[http://parts.igem.org/Part:BBa_J09855 BBa_J09855] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_E0840 BBa_E0840] insert (XbaI, PstI) |
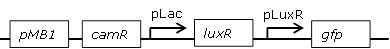 |
| 2 |
Library of the Receiver cell constructs |
Using the BBa_J09855-E0840 construct a library with mutated pLux promoters was created through site-saturation mutagenesis to screen for promoters with changed sensitivities for OHHL: |
 |
| 3 |
Receiver cell construct for GFP experiments without the LuxR generating part |
[http://parts.igem.org/Part:BBa_R0062 BBa_R0062] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_E0840 BBa_E0840] insert (XbaI, PstI) |

 |
| LuxI generating constructs |
| |
Description |
Cloning |
Maps |
| 4 |
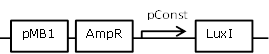
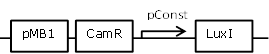
Sender cell construct with a very strong constitutive promoter from the BBa_J23100 promoter library for GFP and Hydrolase experiments |
[http://parts.igem.org/Part:BBa_J23100 BBa_J23100] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_K805016 BBa_K805016] insert (XbaI, PstI) |
 |
| 5 |
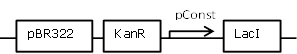
Sender cell construct with an intermediate constitutive promoter from the BBa_J23100 promoter library for GFP and Hydrolase experiments |
[http://parts.igem.org/Part:BBa_J23118 BBa_J23118] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_K805016 BBa_K805016] insert (XbaI, PstI) |


 |
| 6 |
Sender cell construct with an intermediate constitutive promoter from the BBa_J23100 promoter library for GFP and Hydrolase experiments |
[http://parts.igem.org/Part:BBa_J23110 BBa_J23110] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_K805016 BBa_K805016] insert (XbaI, PstI) |


 |
| 7 |
Sender cell construct with a weak constitutive promoter from the BBa_J23100 promoter library for GFP and Hydrolase experiments |
[http://parts.igem.org/Part:BBa_J23114 BBa_J23114] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_K805016 BBa_K805016] insert (XbaI, PstI) |

 |
| LuxR generating constructs |
| |
Description |
Cloning |
Maps |
| 8 |
constitutive LuxR generating biobrick |
[http://parts.igem.org/Part:BBa_J09855 BBa_J09855] |
 |
| 9 |
constitutive LuxR generating biobrick, with negative feedback-loop at high OHHL concentrations |
[http://parts.igem.org/Part:BBa_F2621 BBa_F2621] |
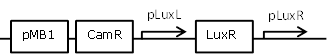 |
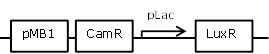
| 10 |
constitutive LuxR generating construct, repressible through LacI |
[http://parts.igem.org/Part:BBa_R0010 BBa_R0010] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_I0462 BBa_I0462] insert (XbaI, PstI) |
 |
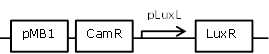
| 11 |
constitutive LuxR generating construct, with negative feedback-loop at high OHHL concentrations |
[http://parts.igem.org/Part:BBa_R0063 BBa_R0063] backbone (SpeI,PstI) and [http://parts.igem.org/Part:BBa_I0462 BBa_I0462] insert (XbaI, PstI) |
 |
| 12 |
auto-inducible LuxR generating construct with positive feedback loop |
[http://parts.igem.org/Part:BBa_R0062 BBa_R0062] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_I0462 BBa_I0462] insert (XbaI, PstI) |
 |
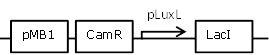
| 13 |
negatively regulated pLuxL-LacI construct to improve leakiness of LuxR system |
[http://parts.igem.org/Part:BBa_R0063 BBa_R0063] backbone (SpeI, PstI) and [http://parts.igem.org/Part:BBa_C0012 BBa_C0012] insert (SpeI,PstI) |
 |
| pLux constructs |
| |
Description |
Cloning |
Maps |
| |
|
|
|
| Hydrolase constructs |
| |
Description |
Cloning |
Maps |
| |
|
|
|
 "
"






