Team:DTU-Denmark/Notebook/19 August 2013
From 2013.igem.org
(→Preparation of constructs to send for sequencing) |
(→Lab 208) |
||
| (29 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:DTU-Denmark/Templates/StartPage|19 August 2013}} | {{:Team:DTU-Denmark/Templates/StartPage|19 August 2013}} | ||
| - | + | Navigate to the [[Team:DTU-Denmark/Notebook/16_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/20_August_2013|Next]] Entry | |
| - | = | + | =Lab 208= |
<hr/> | <hr/> | ||
==Main purpose== | ==Main purpose== | ||
<hr/> | <hr/> | ||
| + | *Preparation of constructs to send for sequencing | ||
| + | *Gradient PCR for creation of Biobricks | ||
| + | *PCR to linearize pZA21::RFP::araBAD | ||
| + | *make Nir1 and Nir2 templates for Morten Nørholm | ||
| + | *check-up PCR of Hello World construct | ||
==Who was in the lab== | ==Who was in the lab== | ||
| Line 15: | Line 20: | ||
===Preparation of constructs to send for sequencing=== | ===Preparation of constructs to send for sequencing=== | ||
| - | Sec2, TAT3-2, TAT3-1a, CycAX and HAO constructs were prepared for sequencing. | + | Sec2, TAT3-2, TAT3-1a, CycAX and HAO constructs were prepared for sequencing. Sample preparation was performed following the indications given by the EZ-Seq service by Macrogen: |
| - | + | ||
| - | [[File:Construct for sequencing - Sheet1.pdf]] | + | 5uL of 100~150ng/ul plasmid + 5uL of 5uM primers. |
| + | |||
| + | An overview of the sample composition can be found in this | ||
| + | [https://static.igem.org/mediawiki/2013/9/90/Construct_for_sequencing_-_Sheet1.pdf file] | ||
| + | <!-- [[File:Construct for sequencing - Sheet1.pdf]] --> | ||
| + | |||
| + | The samples were sent to Macrogen for sequencing. | ||
===Gradient PCR for creation of Biobricks=== | ===Gradient PCR for creation of Biobricks=== | ||
Made a screening to find good conditions for the extraction of our constructs for subsequent ligation into BioBricks. | Made a screening to find good conditions for the extraction of our constructs for subsequent ligation into BioBricks. | ||
| + | |||
| + | <u>conditions</u>: | ||
| + | |||
| + | A: | ||
| + | * HF buffer | ||
| + | * 2% DMSO | ||
| + | * 1uL 50mM MgCl2 | ||
| + | |||
| + | B: | ||
| + | * HF buffer | ||
| + | * 3% DMSO | ||
| + | * 2uL 50mM MgCl2 | ||
| + | |||
| + | C: | ||
| + | * HF buffer | ||
| + | * 5% DMSO | ||
| + | * 1uL 50mM MgCl2 | ||
| + | |||
| + | D: | ||
| + | * HF buffer | ||
| + | * 3% DMSO | ||
| + | * 1uL 50mM MgCl2 | ||
| + | |||
| + | program: | ||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! temperature !! time !! cycles | ||
| + | |- | ||
| + | | 98C || 2:00 || - | ||
| + | |- | ||
| + | | 98C || 0:10 || 36 | ||
| + | |- | ||
| + | | annealing temp. || 1:00 || 36 | ||
| + | |- | ||
| + | | 72C || 1:00 || 36 | ||
| + | |- | ||
| + | | 72C || 5:00 || - | ||
| + | |- | ||
| + | | 10C || hold || - | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | The annealing temperature was a gradient from 48C to 63C in 12 steps. | ||
| + | {| class="wikitable" style="text-align: right" | ||
| + | |- | ||
| + | | sample # || 1 || 2 || 3 || 4 || 5 || 6 || 7 || 8 || 9 || 10 || 11 || 12 | ||
| + | |- | ||
| + | | temperature || 48.3 || 48.7 || 50.0 || 51.8 || 53.3 || 54.7 || 56.3 || 57.5 || 59.1 || 60.8 || 62.5 || 63 | ||
| + | |} | ||
===PCR to linearize pZA21::RFP::araBAD=== | ===PCR to linearize pZA21::RFP::araBAD=== | ||
| - | + | Used GC buffer and 5% DMSO. Made three samples with varying concentrations of 50mM MgCl2: 0,5 uL, 1uL and 2 uL. | |
| - | Performed | + | template: pZA21::RFP::araBAD |
| + | |||
| + | primers: 55a, 55b | ||
| + | |||
| + | program: | ||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! temperature !! time !! cycles | ||
| + | |- | ||
| + | | 98C || 2:00 || - | ||
| + | |- | ||
| + | | 98C || 0:10 || 36 | ||
| + | |- | ||
| + | | 65C || 1:00 || 36 | ||
| + | |- | ||
| + | | 72C || 3:00 || 36 | ||
| + | |- | ||
| + | | 72C || 5:00 || - | ||
| + | |- | ||
| + | | 10C || hold || - | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ===PCR on one of the Hello World constructs to confirm it's intact=== | ||
| + | |||
| + | Performed PCR on the construct containing Sec2 to check the template is correct because our PCRs on it are not working. | ||
| + | |||
| + | template: Sec2 construct | ||
| + | |||
| + | primers: sequencing primers 4_FW_pZA21 and 5_RV_pZA21 | ||
| + | |||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! temperature !! time !! cycles | ||
| + | |- | ||
| + | | 98C || 10:00 || - | ||
| + | |- | ||
| + | | 98C || 0:10 || 36 | ||
| + | |- | ||
| + | | 61C || 0:20 || 36 | ||
| + | |- | ||
| + | | 72C || 2:00 || 36 | ||
| + | |- | ||
| + | | 72C || 5:00 || - | ||
| + | |- | ||
| + | | 10C || hold || - | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | ===PCR on NIR1 and NIR2 via. colony PCR for template to Morten Nørholm=== | ||
| + | |||
| + | {| class="wikitable" style="text-align: right" | ||
| + | |- | ||
| + | | sample N1 or N2# || HF || HF 2% DMSO 1mL MgCl2 50mM ||HF 5% DMSO || HF 1M Betaine || GC || GC 2% DMSO || GC 5% DMSO || GC 5% DMSO 1mL MgCl2 50mM || GC 5% DMSO 2mL MgCl2 50mM || GC 1M Betaine | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| class="wikitable" style="text-align: right" | ||
| + | ! temperature !! time !! cycles | ||
| + | |- | ||
| + | | 98C || 10:00 || - | ||
| + | |- | ||
| + | | 98C || 0:20 || 14 | ||
| + | |- | ||
| + | | 71C -0.5C increment || 0:30 || 14 | ||
| + | |- | ||
| + | | 72C || 2:30 || 14 | ||
| + | |- | ||
| + | | 98C || 0:20 || 20 | ||
| + | |- | ||
| + | | 64C || 0:30 || 20 | ||
| + | |- | ||
| + | | 72C || 2:30 || 20 | ||
| + | |- | ||
| + | | 72C || 5:00 || - | ||
| + | |- | ||
| + | | 10C || hold || - | ||
| + | |- | ||
| + | |} | ||
==Results== | ==Results== | ||
| Line 35: | Line 170: | ||
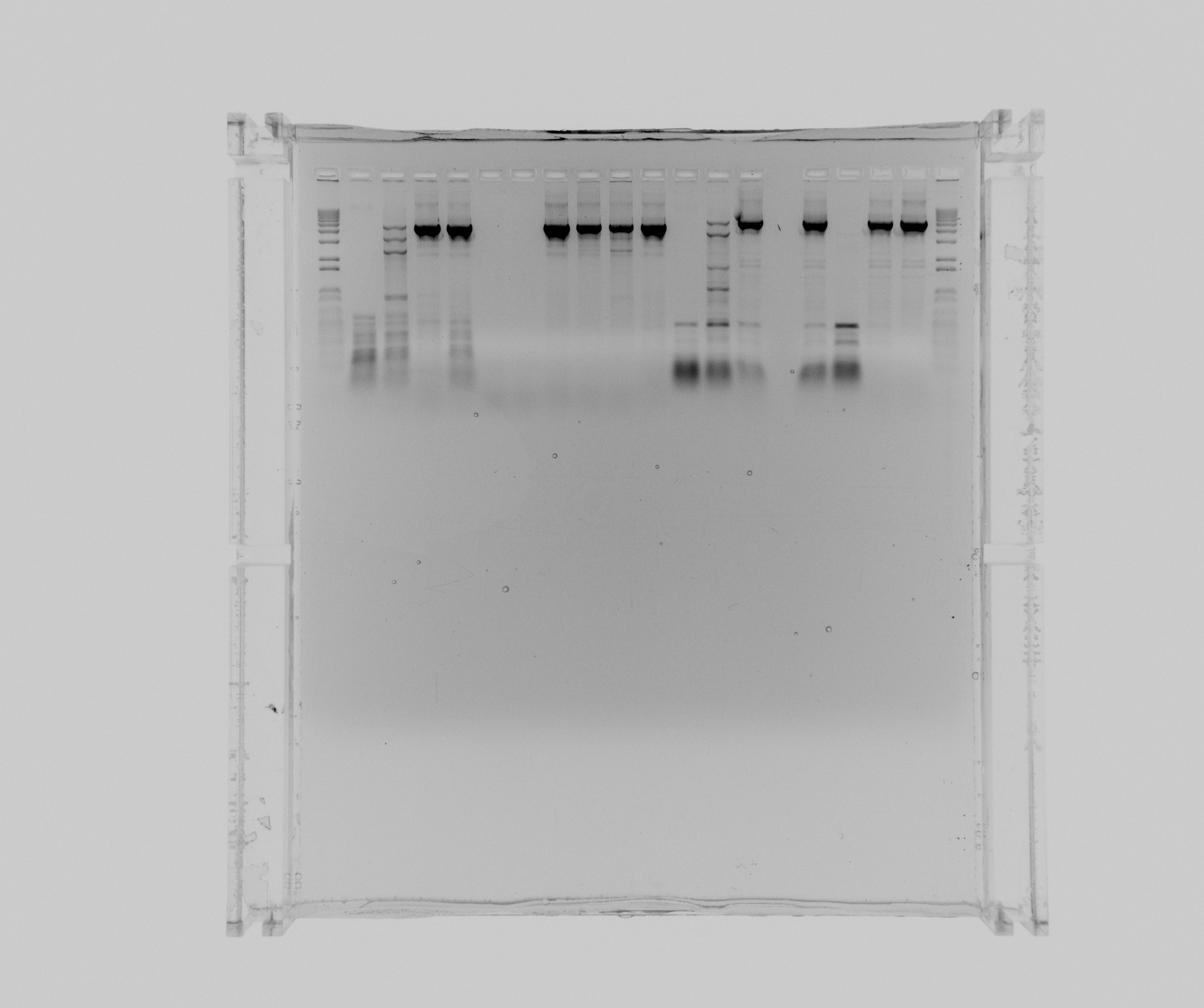
===gels=== | ===gels=== | ||
| - | * 1 kb ladder | + | *1: 1 kb ladder |
| - | * N1 HF | + | *2: N1 HF |
| - | * N1 HF 2% DMSO 1 | + | *3: N1 HF 2% DMSO 1 mL MgCl2 |
| - | * N1 HF 5% DMSO | + | *4: N1 HF 5% DMSO |
| - | * N1 HF | + | *5: N1 HF 1M Betaine |
| - | * N1 GC | + | *6: N1 GC |
| - | * N1 GC 2% DMSO | + | *7: N1 GC 2% DMSO |
| - | * N1 GC 5% DMSO | + | *8: N1 GC 5% DMSO |
| - | * N1 GC 5% DMSO 1 | + | *9: N1 GC 5% DMSO 1 mL MgCL2 |
| - | * N1 GC 5% DMSO 2 | + | *10: N1 GC 5% DMSO 2 mL MgCl2 |
| - | * N1 GC | + | *11: N1 GC 1M Betaine |
| - | * N2 HF | + | *12: N2 HF |
| - | * N2 HF 2% DMSO 1 | + | *13: N2 HF 2% DMSO 1 mL MgCl2 |
| - | * N2 HF 5% DMSO | + | *14: N2 HF 5% DMSO |
| - | * N2 HF | + | *15: N2 HF 1M Betaine |
| - | * N2 GC | + | *16: N2 GC |
| - | * N2 GC 2% DMSO | + | *17: N2 GC 2% DMSO |
| - | * N2 GC 5% DMSO | + | *18: N2 GC 5% DMSO |
| - | * 1 kb ladder | + | *19: 1 kb ladder |
| + | [[File:2013-08-19 big gel.jpg| 600px]] | ||
| + | |||
| - | |||
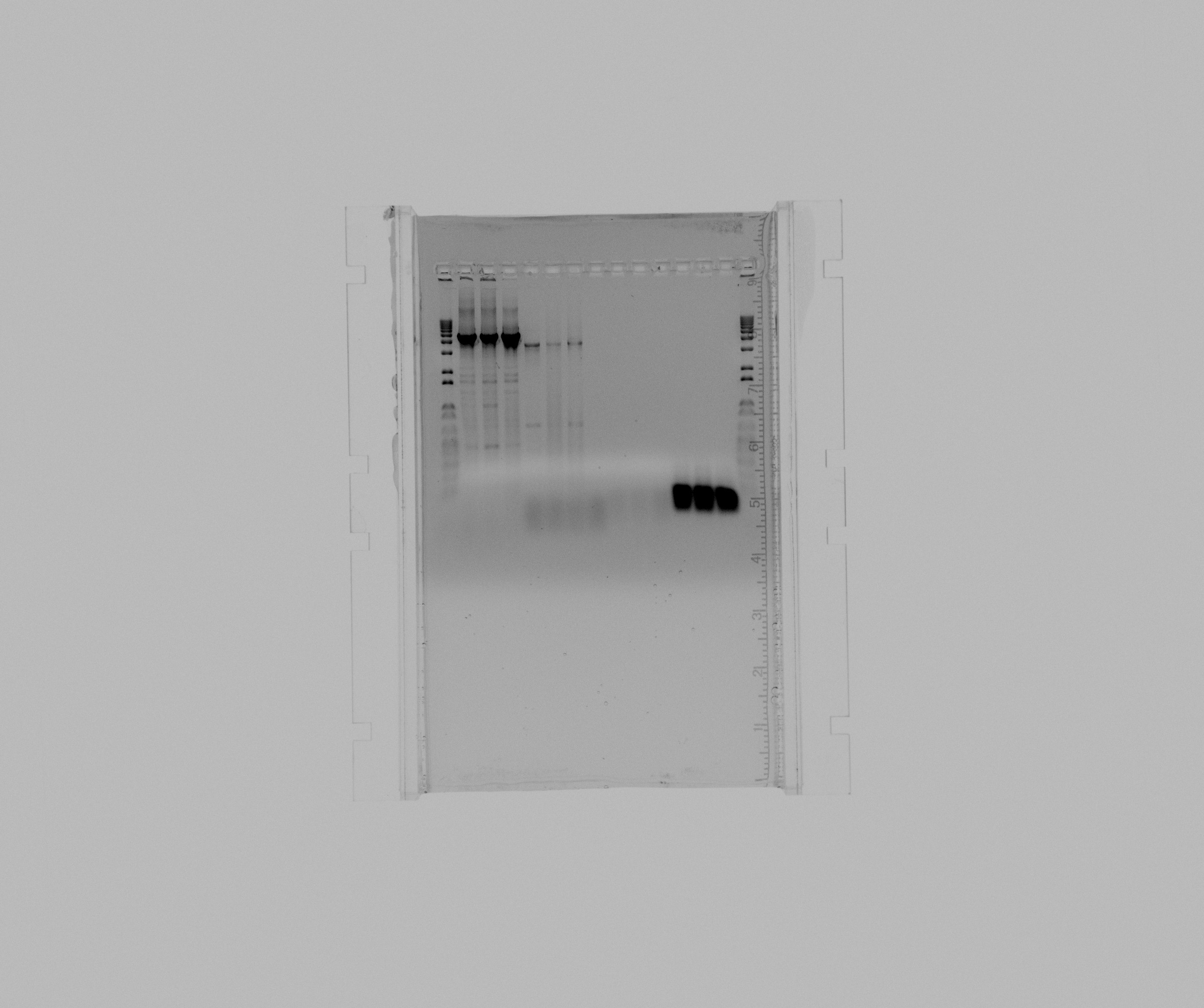
* 1 kb ladder | * 1 kb ladder | ||
| - | * N2 GC 5% DMSO 1 | + | * N2 GC 5% DMSO 1 mL MgCl2 |
| - | * N2 GC 5% DMSO 2 | + | * N2 GC 5% DMSO 2 mL MgCl2 |
| - | * N2 GC | + | * N2 GC 1M Betaine |
* pZA21::RFP::araBAD 0,5 uL MgCl2 | * pZA21::RFP::araBAD 0,5 uL MgCl2 | ||
* pZA21::RFP::araBAD 1 uL MgCl2 | * pZA21::RFP::araBAD 1 uL MgCl2 | ||
* pZA21::RFP::araBAD 2 uL MgCl2 | * pZA21::RFP::araBAD 2 uL MgCl2 | ||
* negative pZA21::RFP::araBAD | * negative pZA21::RFP::araBAD | ||
| - | * | + | * check Sec2 |
| - | * | + | * check Sec2 (duplicate) |
| - | * negative | + | * negative check Sec2 |
* screening A1 | * screening A1 | ||
* screening A2 | * screening A2 | ||
* screening A3 | * screening A3 | ||
| - | + | [[File:2013-08-19 small gel.jpg| 600px]] | |
| - | + | ||
==Conclusion== | ==Conclusion== | ||
<hr/> | <hr/> | ||
| - | Navigate to the [[Team:DTU-Denmark/Notebook/ | + | The program for Nir colony PCR works as a touchdown PCR and it have a big influence if you add DMSO or Betaine. It seems as 5% DMSO and no additional MgCl2 or 1M Betaine works best and somewhat equally good for both reactions. |
| + | |||
| + | =lab 115= | ||
| + | <hr/> | ||
| + | ==Main purpose== | ||
| + | Prepare Nitrate and Nitrite standard solutions for colorimetric analysis | ||
| + | |||
| + | *12 mg/L NO<sub>3</sub>-N | ||
| + | *0.5 mg/L NO<sub>2</sub>-N | ||
| + | |||
| + | ==Who was in the lab== | ||
| + | <hr/> | ||
| + | Ariadni, Helen | ||
| + | |||
| + | ==Procedure== | ||
| + | <hr/> | ||
| + | * Weigh 77.14 mg NaNO<sub>3</sub> and dissolve in 100 ml for 10X solution of nitrate. | ||
| + | * Weigh 123.2 mg NaNO<sub>2</sub> and dissolve in 50 ml for 1000X solution and then dilute 1 ml in 99 ml for 10X solution | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Navigate to the [[Team:DTU-Denmark/Notebook/16_August_2013|Previous]] or the [[Team:DTU-Denmark/Notebook/20_August_2013|Next]] Entry | ||
{{:Team:DTU-Denmark/Templates/EndPage}} | {{:Team:DTU-Denmark/Templates/EndPage}} | ||
Latest revision as of 17:38, 28 September 2013
19 August 2013
Contents |
Lab 208
Main purpose
- Preparation of constructs to send for sequencing
- Gradient PCR for creation of Biobricks
- PCR to linearize pZA21::RFP::araBAD
- make Nir1 and Nir2 templates for Morten Nørholm
- check-up PCR of Hello World construct
Who was in the lab
Kristian, Henrike, Julia
Procedure
Preparation of constructs to send for sequencing
Sec2, TAT3-2, TAT3-1a, CycAX and HAO constructs were prepared for sequencing. Sample preparation was performed following the indications given by the EZ-Seq service by Macrogen:
5uL of 100~150ng/ul plasmid + 5uL of 5uM primers.
An overview of the sample composition can be found in this file
The samples were sent to Macrogen for sequencing.
Gradient PCR for creation of Biobricks
Made a screening to find good conditions for the extraction of our constructs for subsequent ligation into BioBricks.
conditions:
A:
- HF buffer
- 2% DMSO
- 1uL 50mM MgCl2
B:
- HF buffer
- 3% DMSO
- 2uL 50mM MgCl2
C:
- HF buffer
- 5% DMSO
- 1uL 50mM MgCl2
D:
- HF buffer
- 3% DMSO
- 1uL 50mM MgCl2
program:
| temperature | time | cycles |
|---|---|---|
| 98C | 2:00 | - |
| 98C | 0:10 | 36 |
| annealing temp. | 1:00 | 36 |
| 72C | 1:00 | 36 |
| 72C | 5:00 | - |
| 10C | hold | - |
The annealing temperature was a gradient from 48C to 63C in 12 steps.
| sample # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| temperature | 48.3 | 48.7 | 50.0 | 51.8 | 53.3 | 54.7 | 56.3 | 57.5 | 59.1 | 60.8 | 62.5 | 63 |
PCR to linearize pZA21::RFP::araBAD
Used GC buffer and 5% DMSO. Made three samples with varying concentrations of 50mM MgCl2: 0,5 uL, 1uL and 2 uL.
template: pZA21::RFP::araBAD
primers: 55a, 55b
program:
| temperature | time | cycles |
|---|---|---|
| 98C | 2:00 | - |
| 98C | 0:10 | 36 |
| 65C | 1:00 | 36 |
| 72C | 3:00 | 36 |
| 72C | 5:00 | - |
| 10C | hold | - |
PCR on one of the Hello World constructs to confirm it's intact
Performed PCR on the construct containing Sec2 to check the template is correct because our PCRs on it are not working.
template: Sec2 construct
primers: sequencing primers 4_FW_pZA21 and 5_RV_pZA21
| temperature | time | cycles |
|---|---|---|
| 98C | 10:00 | - |
| 98C | 0:10 | 36 |
| 61C | 0:20 | 36 |
| 72C | 2:00 | 36 |
| 72C | 5:00 | - |
| 10C | hold | - |
PCR on NIR1 and NIR2 via. colony PCR for template to Morten Nørholm
| sample N1 or N2# | HF | HF 2% DMSO 1mL MgCl2 50mM | HF 5% DMSO | HF 1M Betaine | GC | GC 2% DMSO | GC 5% DMSO | GC 5% DMSO 1mL MgCl2 50mM | GC 5% DMSO 2mL MgCl2 50mM | GC 1M Betaine |
| temperature | time | cycles |
|---|---|---|
| 98C | 10:00 | - |
| 98C | 0:20 | 14 |
| 71C -0.5C increment | 0:30 | 14 |
| 72C | 2:30 | 14 |
| 98C | 0:20 | 20 |
| 64C | 0:30 | 20 |
| 72C | 2:30 | 20 |
| 72C | 5:00 | - |
| 10C | hold | - |
Results
gels
- 1: 1 kb ladder
- 2: N1 HF
- 3: N1 HF 2% DMSO 1 mL MgCl2
- 4: N1 HF 5% DMSO
- 5: N1 HF 1M Betaine
- 6: N1 GC
- 7: N1 GC 2% DMSO
- 8: N1 GC 5% DMSO
- 9: N1 GC 5% DMSO 1 mL MgCL2
- 10: N1 GC 5% DMSO 2 mL MgCl2
- 11: N1 GC 1M Betaine
- 12: N2 HF
- 13: N2 HF 2% DMSO 1 mL MgCl2
- 14: N2 HF 5% DMSO
- 15: N2 HF 1M Betaine
- 16: N2 GC
- 17: N2 GC 2% DMSO
- 18: N2 GC 5% DMSO
- 19: 1 kb ladder
- 1 kb ladder
- N2 GC 5% DMSO 1 mL MgCl2
- N2 GC 5% DMSO 2 mL MgCl2
- N2 GC 1M Betaine
- pZA21::RFP::araBAD 0,5 uL MgCl2
- pZA21::RFP::araBAD 1 uL MgCl2
- pZA21::RFP::araBAD 2 uL MgCl2
- negative pZA21::RFP::araBAD
- check Sec2
- check Sec2 (duplicate)
- negative check Sec2
- screening A1
- screening A2
- screening A3
Conclusion
The program for Nir colony PCR works as a touchdown PCR and it have a big influence if you add DMSO or Betaine. It seems as 5% DMSO and no additional MgCl2 or 1M Betaine works best and somewhat equally good for both reactions.
lab 115
Main purpose
Prepare Nitrate and Nitrite standard solutions for colorimetric analysis
- 12 mg/L NO3-N
- 0.5 mg/L NO2-N
Who was in the lab
Ariadni, Helen
Procedure
- Weigh 77.14 mg NaNO3 and dissolve in 100 ml for 10X solution of nitrate.
- Weigh 123.2 mg NaNO2 and dissolve in 50 ml for 1000X solution and then dilute 1 ml in 99 ml for 10X solution
Navigate to the Previous or the Next Entry
 "
"